Protein-DNA interaction mapping using genomic tiling path microarrays in Drosophila
- PMID: 12876199
- PMCID: PMC170935
- DOI: 10.1073/pnas.1533393100
Protein-DNA interaction mapping using genomic tiling path microarrays in Drosophila
Abstract
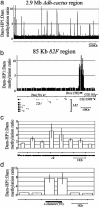
We demonstrate the use of a chromosomal walk (or "tiling path") printed as DNA microarrays for mapping protein-DNA interactions across large regions of contiguous genomic DNA in Drosophila melanogaster. Microarrays were constructed with genomic DNA fragments 430-920 bp in length, covering 2.9 million base pairs of the Adh-cactus region of chromosome 2 and 85,000 base pairs of the 82F region of chromosome 3. We performed DNA localization mapping for the heterochromatin protein HP1 and for the sequence-specific GAGA transcription factor, producing a comprehensive, high-resolution map of in vivo protein-DNA interactions throughout these regions of the Drosophila genome.
Figures


References
-
- Reinke, V. & White, K. P. (2002) Annu. Rev. Genomics Hum. Genet. 3, 153–178. - PubMed
-
- White, K. P., Rifkin, S. A., Hurban, P. & Hogness, D. S. (1999) Science 286, 2179–2184. - PubMed
-
- Reinke, V., Smith, H. E., Nance, J., Wang, J., Van Doren, C., Begley, R., Jones, S. J., Davis, E. B., Scherer, S., Ward, S. & Kim, S. K. (2000) Mol. Cell 6, 605–616. - PubMed
-
- Hill, A. A., Hunter, C. P., Tsung, B. T., Tucker-Kellogg, G. & Brown, E. L. (2000) Science 290, 809–812. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

