A novel role for dp115 in the organization of tER sites in Drosophila
- PMID: 12876273
- PMCID: PMC2172793
- DOI: 10.1083/jcb.200301136
A novel role for dp115 in the organization of tER sites in Drosophila
Abstract
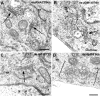
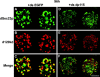
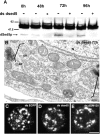
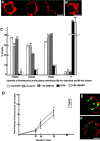
Here, we describe that depletion of the Drosophila homologue of p115 (dp115) by RNA interference in Drosophila S2 cells led to important morphological changes in the Golgi stack morphology and the transitional ER (tER) organization. Using conventional and immunoelectron microscopy and confocal immunofluorescence microscopy, we show that Golgi stacks were converted into clusters of vesicles and tubules, and that the tERs (marked by Sec23p) lost their focused organization and were now dispersed throughout the cytoplasm. However, we found that this morphologically altered exocytic pathway was nevertheless largely competent in anterograde protein transport using two different assays. The effects were specific for dp115. Depletion of the Drosophila homologues of GM130 and syntaxin 5 (dSed5p) did not lead to an effect on the tER organization, though the Golgi stacks were greatly vesiculated in the cells depleted of dSed5p. Taken together, these studies suggest that dp115 could be implicated in the architecture of both the Golgi stacks and the tER sites.
Figures





References
-
- Allan, B.B., B.D. Moyer, and W.E. Balch. 2000. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 289:444–448. - PubMed
-
- Alvarez, C., R. Garcia-Mata, H.P. Hauri, and E. Sztul. 2001. The p115-interactive proteins GM130 and giantin participate in endoplasmic reticulum-Golgi traffic. J. Biol. Chem. 276:2693–2700. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

