Dual function of Slit2 in repulsion and enhanced migration of trunk, but not vagal, neural crest cells
- PMID: 12876276
- PMCID: PMC2172792
- DOI: 10.1083/jcb.200301041
Dual function of Slit2 in repulsion and enhanced migration of trunk, but not vagal, neural crest cells
Abstract
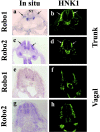
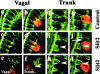
Neural crest precursors to the autonomic nervous system form different derivatives depending upon their axial level of origin; for example, vagal, but not trunk, neural crest cells form the enteric ganglia of the gut. Here, we show that Slit2 is expressed at the entrance of the gut, which is selectively invaded by vagal, but not trunk, neural crest. Accordingly, only trunk neural crest cells express Robo receptors. In vivo and in vitro experiments demonstrate that trunk, not vagal, crest cells avoid cells or cell membranes expressing Slit2, thereby contributing to the differential ability of neural crest populations to invade and innervate the gut. Conversely, exposure to soluble Slit2 significantly increases the distance traversed by trunk neural crest cells. These results suggest that Slit2 can act bifunctionally, both repulsing and stimulating the motility of trunk neural crest cells.
Figures





Similar articles
-
Sacral neural crest cells colonise aganglionic hindgut in vivo but fail to compensate for lack of enteric ganglia.Dev Biol. 2000 Mar 1;219(1):30-43. doi: 10.1006/dbio.1999.9592. Dev Biol. 2000. PMID: 10677253
-
Slit molecules prevent entrance of trunk neural crest cells in developing gut.Int J Dev Neurosci. 2015 Apr;41:8-16. doi: 10.1016/j.ijdevneu.2014.12.003. Epub 2014 Dec 6. Int J Dev Neurosci. 2015. PMID: 25490618 Free PMC article.
-
Colonization of the bowel by neural crest-derived cells re-migrating from foregut backtransplanted to vagal or sacral regions of host embryos.Dev Dyn. 1993 Mar;196(3):217-33. doi: 10.1002/aja.1001960308. Dev Dyn. 1993. PMID: 8400406
-
Guidance cues involved in the development of the peripheral autonomic nervous system.Auton Neurosci. 2004 May 31;112(1-2):1-14. doi: 10.1016/j.autneu.2004.02.008. Auton Neurosci. 2004. PMID: 15233925 Review.
-
Migration and diversification of the vagal neural crest.Dev Biol. 2018 Dec 1;444 Suppl 1(Suppl 1):S98-S109. doi: 10.1016/j.ydbio.2018.07.004. Epub 2018 Jul 5. Dev Biol. 2018. PMID: 29981692 Free PMC article. Review.
Cited by
-
Roundabout receptors are critical for foregut separation from the body wall.Dev Cell. 2013 Jan 14;24(1):52-63. doi: 10.1016/j.devcel.2012.11.018. Dev Cell. 2013. PMID: 23328398 Free PMC article.
-
Modelling collective cell migration of neural crest.Curr Opin Cell Biol. 2016 Oct;42:22-28. doi: 10.1016/j.ceb.2016.03.023. Epub 2016 Apr 13. Curr Opin Cell Biol. 2016. PMID: 27085004 Free PMC article. Review.
-
Slits affect the timely migration of neural crest cells via Robo receptor.Dev Dyn. 2012 Aug;241(8):1274-88. doi: 10.1002/dvdy.23817. Epub 2012 Jun 23. Dev Dyn. 2012. PMID: 22689303 Free PMC article.
-
Neuregulin-1 is a chemoattractant and chemokinetic molecule for trunk neural crest cells.Dev Dyn. 2018 Jul;247(7):888-902. doi: 10.1002/dvdy.24625. Epub 2018 Mar 25. Dev Dyn. 2018. PMID: 29516589 Free PMC article.
-
The chemokine stromal cell-derived factor-1 regulates the migration of sensory neuron progenitors.J Neurosci. 2005 Apr 20;25(16):3995-4003. doi: 10.1523/JNEUROSCI.4631-04.2005. J Neurosci. 2005. PMID: 15843601 Free PMC article.
References
-
- Bagri, A., O. Marin, A.S. Plump, J. Mak, S.J. Pleasure, J.L. Rubenstein, and M. Tessier-Lavigne. 2002. Slit proteins prevent midline crossing and determine the dorsoventral position of major axonal pathways in the mammalian forebrain. Neuron. 33:233–248. - PubMed
-
- Bronner-Fraser, M. 1982. Analysis of neural crest migration and differentiation using a microinjection technique. Int. J. Neurol. 16-17:73–94. - PubMed
-
- Bronner-Fraser, M., and A.M. Cohen. 1980. Analysis of the neural crest ventral pathway using injected tracer cells. Dev. Biol. 77:130–141. - PubMed
-
- Bronner-Fraser, M., C.D. Stern, and S. Fraser. 1991. Analysis of neural crest cell lineage and migration. J. Craniofac. Genet. Dev. Biol. 11:214–222. - PubMed

