Phylogenetic analysis of bacterial and archaeal arsC gene sequences suggests an ancient, common origin for arsenate reductase
- PMID: 12877744
- PMCID: PMC183826
- DOI: 10.1186/1471-2148-3-18
Phylogenetic analysis of bacterial and archaeal arsC gene sequences suggests an ancient, common origin for arsenate reductase
Abstract
Background: The ars gene system provides arsenic resistance for a variety of microorganisms and can be chromosomal or plasmid-borne. The arsC gene, which codes for an arsenate reductase is essential for arsenate resistance and transforms arsenate into arsenite, which is extruded from the cell. A survey of GenBank shows that arsC appears to be phylogenetically widespread both in organisms with known arsenic resistance and those organisms that have been sequenced as part of whole genome projects.
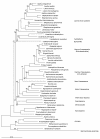
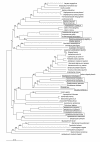
Results: Phylogenetic analysis of aligned arsC sequences shows broad similarities to the established 16S rRNA phylogeny, with separation of bacterial, archaeal, and subsequently eukaryotic arsC genes. However, inconsistencies between arsC and 16S rRNA are apparent for some taxa. Cyanobacteria and some of the gamma-Proteobacteria appear to possess arsC genes that are similar to those of Low GC Gram-positive Bacteria, and other isolated taxa possess arsC genes that would not be expected based on known evolutionary relationships. There is no clear separation of plasmid-borne and chromosomal arsC genes, although a number of the Enterobacteriales (gamma-Proteobacteria) possess similar plasmid-encoded arsC sequences.
Conclusion: The overall phylogeny of the arsenate reductases suggests a single, early origin of the arsC gene and subsequent sequence divergence to give the distinct arsC classes that exist today. Discrepancies between 16S rRNA and arsC phylogenies support the role of horizontal gene transfer (HGT) in the evolution of arsenate reductases, with a number of instances of HGT early in bacterial arsC evolution. Plasmid-borne arsC genes are not monophyletic suggesting multiple cases of chromosomal-plasmid exchange and subsequent HGT. Overall, arsC phylogeny is complex and is likely the result of a number of evolutionary mechanisms.
Figures

References
-
- Tamaki S, Frankenberger WT., Jr Environmental biochemistry of arsenic. Rev Environ Cont Toxicol. 1992;124:79–110. - PubMed
-
- Anderson GL, Williams J, Hille R. The purification and characterization of arsenite oxidase from Alcaligenes faecalis, a molybdenum-containing hydroxylase. J Biol Chem. 1992;267:23674–23682. - PubMed
-
- Abdrashitova SA, Abdullina GG, Ilyaletdinov AN. Role of arsenites in lipid peroxidation in Pseudomonas putida cells oxidizing arsenite. Mikrobiologiya. 1986;55:212–216.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

