Efficient RNA 2'-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C'/D' RNPs
- PMID: 12881427
- PMCID: PMC169041
- DOI: 10.1093/emboj/cdg368
Efficient RNA 2'-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C'/D' RNPs
Abstract
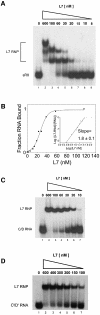
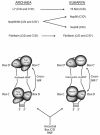
Box C/D ribonucleoprotein (RNP) complexes direct the nucleotide-specific 2'-O-methylation of ribonucleotide sugars in target RNAs. In vitro assembly of an archaeal box C/D sRNP using recombinant core proteins L7, Nop56/58 and fibrillarin has yielded an RNA:protein enzyme that guides methylation from both the terminal box C/D core and internal C'/D' RNP complexes. Reconstitution of sRNP complexes containing only box C/D or C'/D' motifs has demonstrated that the terminal box C/D RNP is the minimal methylation-competent particle. However, efficient ribonucleotide 2'-O-methylation requires that both the box C/D and C'/D' RNPs function within the full-length sRNA molecule. In contrast to the eukaryotic snoRNP complex, where the core proteins are distributed asymmetrically on the box C/D and C'/D' motifs, all three archaeal core proteins bind both motifs symmetrically. This difference in core protein distribution is a result of altered RNA-binding capabilities of the archaeal and eukaryotic core protein homologs. Thus, evolution of the box C/D nucleotide modification complex has resulted in structurally distinct archaeal and eukaryotic RNP particles.
Figures




Similar articles
-
Conserved spacing between the box C/D and C'/D' RNPs of the archaeal box C/D sRNP complex is required for efficient 2'-O-methylation of target RNAs.RNA. 2005 Mar;11(3):285-93. doi: 10.1261/rna.7223405. Epub 2005 Jan 20. RNA. 2005. PMID: 15661846 Free PMC article.
-
The coiled-coil domain of the Nop56/58 core protein is dispensable for sRNP assembly but is critical for archaeal box C/D sRNP-guided nucleotide methylation.RNA. 2006 Jun;12(6):1092-103. doi: 10.1261/rna.2230106. Epub 2006 Apr 6. RNA. 2006. PMID: 16601205 Free PMC article.
-
Assembly of the archaeal box C/D sRNP can occur via alternative pathways and requires temperature-facilitated sRNA remodeling.J Mol Biol. 2006 Oct 6;362(5):1025-42. doi: 10.1016/j.jmb.2006.07.091. Epub 2006 Aug 4. J Mol Biol. 2006. PMID: 16949610
-
In vitro reconstitution and affinity purification of catalytically active archaeal box C/D sRNP complexes.Methods Enzymol. 2007;425:263-82. doi: 10.1016/S0076-6879(07)25012-8. Methods Enzymol. 2007. PMID: 17673088 Review.
-
RNA structure and function in C/D and H/ACA s(no)RNPs.Curr Opin Struct Biol. 2004 Jun;14(3):335-43. doi: 10.1016/j.sbi.2004.05.006. Curr Opin Struct Biol. 2004. PMID: 15193314 Review.
Cited by
-
Structurally conserved Nop56/58 N-terminal domain facilitates archaeal box C/D ribonucleoprotein-guided methyltransferase activity.J Biol Chem. 2012 Jun 1;287(23):19418-28. doi: 10.1074/jbc.M111.323253. Epub 2012 Apr 11. J Biol Chem. 2012. PMID: 22496443 Free PMC article.
-
Illumina-based RiboMethSeq approach for mapping of 2'-O-Me residues in RNA.Nucleic Acids Res. 2016 Sep 19;44(16):e135. doi: 10.1093/nar/gkw547. Epub 2016 Jun 14. Nucleic Acids Res. 2016. PMID: 27302133 Free PMC article.
-
The structure and function of small nucleolar ribonucleoproteins.Nucleic Acids Res. 2007;35(5):1452-64. doi: 10.1093/nar/gkl1172. Epub 2007 Feb 6. Nucleic Acids Res. 2007. PMID: 17284456 Free PMC article. Review.
-
Structural organization of box C/D RNA-guided RNA methyltransferase.Proc Natl Acad Sci U S A. 2009 Aug 18;106(33):13808-13. doi: 10.1073/pnas.0905128106. Epub 2009 Aug 5. Proc Natl Acad Sci U S A. 2009. PMID: 19666563 Free PMC article.
-
Structural features of the guide:target RNA duplex required for archaeal box C/D sRNA-guided nucleotide 2'-O-methylation.RNA. 2007 Jun;13(6):899-911. doi: 10.1261/rna.517307. Epub 2007 Apr 16. RNA. 2007. PMID: 17438123 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

