Interaction between Ski7p and Upf1p is required for nonsense-mediated 3'-to-5' mRNA decay in yeast
- PMID: 12881429
- PMCID: PMC169047
- DOI: 10.1093/emboj/cdg374
Interaction between Ski7p and Upf1p is required for nonsense-mediated 3'-to-5' mRNA decay in yeast
Abstract
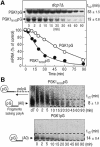
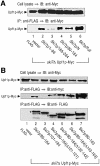
Aberrant mRNAs containing premature termination codons (PTC-mRNAs) are degraded by a conserved surveillance system, referred to as the nonsense- mediated decay (NMD) pathway. Although NMD is reported to operate on the decapping and 5'-to-3' exonucleolytic decay of PTC-mRNAs without affecting deadenylation, a role for an opposite 3'-to-5' decay pathway remains largely unexplored. In this study, we have characterized the 3'-to-5' directed mRNA degradation in the yeast NMD pathway. PTC-mRNAs are stabilized in yeast cells lacking the components of 3'-to-5' mRNA-decay machinery. The 3'-to-5' directed degradation of PTC-mRNAs proceeds more rapidly than that of the PTC-free transcript, in a manner dependent on the cytoplasmic exosome and Upf proteins. Moreover, Upf1p, but not Upf2p, interacts physically with an N-terminal domain of Ski7p, although the interaction requires Upf2p. The efficiency of 3'-to-5' directed degradation of PTC-mRNAs is impaired by overexpression of Ski7p N-domain fragments that contain a sequence of the Upf1p-interaction region. These data suggest that the activation of 3'-to-5' directed NMD is mediated through the interaction between Upf1p and the Ski7p N domain.
Figures






Similar articles
-
An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3'-->5' degradation.Mol Cell. 2003 May;11(5):1405-13. doi: 10.1016/s1097-2765(03)00190-4. Mol Cell. 2003. PMID: 12769863
-
Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover.Mol Cell Biol. 1996 Oct;16(10):5491-506. doi: 10.1128/MCB.16.10.5491. Mol Cell Biol. 1996. PMID: 8816462 Free PMC article.
-
Targeting of aberrant mRNAs to cytoplasmic processing bodies.Cell. 2006 Jun 16;125(6):1095-109. doi: 10.1016/j.cell.2006.04.037. Cell. 2006. PMID: 16777600 Free PMC article.
-
NMD monitors translational fidelity 24/7.Curr Genet. 2017 Dec;63(6):1007-1010. doi: 10.1007/s00294-017-0709-4. Epub 2017 May 23. Curr Genet. 2017. PMID: 28536849 Free PMC article. Review.
-
Cutting the nonsense: the degradation of PTC-containing mRNAs.Biochem Soc Trans. 2010 Dec;38(6):1615-20. doi: 10.1042/BST0381615. Biochem Soc Trans. 2010. PMID: 21118136 Review.
Cited by
-
A Two-Headed Monster to Avert Disaster: HBS1/SKI7 Is Alternatively Spliced to Build Eukaryotic RNA Surveillance Complexes.Front Plant Sci. 2018 Sep 12;9:1333. doi: 10.3389/fpls.2018.01333. eCollection 2018. Front Plant Sci. 2018. PMID: 30258456 Free PMC article.
-
Saccharomyces cerevisiae Ebs1p is a putative ortholog of human Smg7 and promotes nonsense-mediated mRNA decay.Nucleic Acids Res. 2007;35(22):7688-97. doi: 10.1093/nar/gkm912. Epub 2007 Nov 4. Nucleic Acids Res. 2007. PMID: 17984081 Free PMC article.
-
RNA recognition by 3'-to-5' exonucleases: the substrate perspective.Biochim Biophys Acta. 2008 Apr;1779(4):256-65. doi: 10.1016/j.bbagrm.2007.11.004. Epub 2007 Dec 3. Biochim Biophys Acta. 2008. PMID: 18078842 Free PMC article. Review.
-
The role of deadenylation in the degradation of unstable mRNAs in trypanosomes.Nucleic Acids Res. 2009 Sep;37(16):5511-28. doi: 10.1093/nar/gkp571. Epub 2009 Jul 13. Nucleic Acids Res. 2009. PMID: 19596809 Free PMC article.
-
The exosome controls alternative splicing by mediating the gene expression and assembly of the spliceosome complex.Sci Rep. 2015 Aug 26;5:13403. doi: 10.1038/srep13403. Sci Rep. 2015. PMID: 26306464 Free PMC article.
References
-
- Atkin A.L., Schenkman,L.R., Eastham,M., Dahlseid,J.N., Lelivelt,M.J. and Culbertson,M.R. (1997) Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem., 272, 22163–22172. - PubMed
-
- Beelman C.A., Stevens,A., Caponigro,G., LaGrandeur,T.E., Hatfield,L., Fortner,D.M. and Parker,R. (1996) An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature, 382, 642–646. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

