Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B
- PMID: 12883001
- PMCID: PMC170954
- DOI: 10.1073/pnas.1033001100
Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B
Abstract
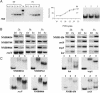
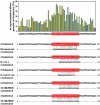
Iron is limiting in the human host, and bacterial pathogens respond to this environment by activating genes required for bacterial virulence. Transcriptional regulation in response to iron in Gram-negative bacteria is largely mediated by the ferric uptake regulator protein Fur, which in the presence of iron binds to a specific sequence in the promoter regions of genes under its control and acts as a repressor. Here we describe DNA microarray, computational and in vitro studies to define the Fur regulon in the human pathogen Neisseria meningitidis group B (strain MC58). After iron addition to an iron-depleted bacterial culture, 153 genes were up-regulated and 80 were down-regulated. Only 50% of the iron-regulated genes were found to contain Fur-binding consensus sequences in their promoter regions. Forty-two promoter regions were amplified and 32 of these were shown to bind Fur by gel-shift analysis. Among these genes, many of which had never been described before to be Fur-regulated, 10 were up-regulated on iron addition, demonstrating that Fur can also act as a transcriptional activator. Sequence alignment of the Fur-binding regions revealed that the N. meningitidis Fur-box encompasses the highly conserved (NATWAT)3 motif. Cluster analysis was effective in predicting Fur-regulated genes even if computer prediction failed to identify Fur-box-like sequences in their promoter regions. Microarray-generated gene expression profiling appears to be a very effective approach to define new regulons and regulatory pathways in pathogenic bacteria.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

