Reverse genetic system for the analysis of parvovirus telomeres reveals interactions between transcription factor binding sites in the hairpin stem
- PMID: 12885883
- PMCID: PMC167226
- DOI: 10.1128/jvi.77.16.8650-8660.2003
Reverse genetic system for the analysis of parvovirus telomeres reveals interactions between transcription factor binding sites in the hairpin stem
Abstract
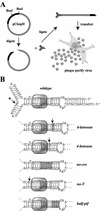
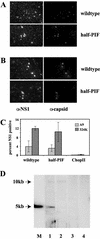
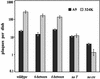
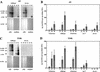
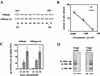
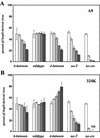
The left-hand or 3'-terminal hairpin of minute virus of mice (MVM) contains sequence elements essential for both viral DNA replication at the left-hand origin (oriL) and for the modulation of the P4 promoter, from which the viral nonstructural gene cassette is transcribed. This hairpin sequence has proven difficult to manipulate in the context of the viral genome. Here we describe a system for generating mutant viruses using synthetic hairpin oligonucleotides and a truncated form of the infectious clone. This allows manipulation of the sequence of the left-hand hairpin and examination of the effects in the context of the viral life cycle. We have confirmed the requirement for a functional parvovirus initiation factor (PIF) binding site and determined that an optimized PIF binding site, with 6 bases between the half-sites, was actually detrimental to viral growth. The distal PIF half-site overlaps a cyclic AMP-responsive element (CRE), which was shown to play an important role in initiating infection, particularly in 324K simian virus 40-transformed human fibroblasts. Interestingly, reducing the spacing of the PIF half-sites, and thus the affinity of the binding site for PIF, increased viral fitness relative to wild type in 324K cells, but not in murine A9 cells. These results indicate that the relative importance of factor binding to the CRE and PIF sites during the establishment of an infection differs markedly between these two host cells and suggest that the suboptimal spacing of PIF half-sites found in wild-type virus represents a necessary reduction in the affinity of the PIF interaction in favor of CRE function.
Figures


Similar articles
-
Segregation of a single outboard left-end origin is essential for the viability of parvovirus minute virus of mice.J Virol. 2006 Nov;80(21):10879-83. doi: 10.1128/JVI.01501-06. Epub 2006 Aug 23. J Virol. 2006. PMID: 16928767 Free PMC article.
-
Exploring the contribution of distal P4 promoter elements to the oncoselectivity of Minute Virus of Mice.Virology. 2007 Apr 25;361(1):174-84. doi: 10.1016/j.virol.2006.11.006. Epub 2006 Dec 18. Virology. 2007. PMID: 17175002 Free PMC article.
-
Minute virus of mice initiator protein NS1 and a host KDWK family transcription factor must form a precise ternary complex with origin DNA for nicking to occur.J Virol. 2001 Aug;75(15):7009-17. doi: 10.1128/JVI.75.15.7009-7017.2001. J Virol. 2001. PMID: 11435581 Free PMC article.
-
Parvovirus diversity and DNA damage responses.Cold Spring Harb Perspect Biol. 2013 Feb 1;5(2):a012989. doi: 10.1101/cshperspect.a012989. Cold Spring Harb Perspect Biol. 2013. PMID: 23293137 Free PMC article. Review.
-
Molecular biology of autonomous parvoviruses.Contrib Microbiol. 2000;4:163-77. doi: 10.1159/000060336. Contrib Microbiol. 2000. PMID: 10941577 Review. No abstract available.
Cited by
-
Maintenance of the flip sequence orientation of the ears in the parvoviral left-end hairpin is a nonessential consequence of the critical asymmetry in the hairpin stem.J Virol. 2012 Nov;86(22):12187-97. doi: 10.1128/JVI.01450-12. Epub 2012 Aug 29. J Virol. 2012. PMID: 22933276 Free PMC article.
-
LuIII parvovirus selectively and efficiently targets, replicates in, and kills human glioma cells.J Virol. 2012 Jul;86(13):7280-91. doi: 10.1128/JVI.00227-12. Epub 2012 May 2. J Virol. 2012. PMID: 22553327 Free PMC article.
-
Segregation of a single outboard left-end origin is essential for the viability of parvovirus minute virus of mice.J Virol. 2006 Nov;80(21):10879-83. doi: 10.1128/JVI.01501-06. Epub 2006 Aug 23. J Virol. 2006. PMID: 16928767 Free PMC article.
-
Exploring the contribution of distal P4 promoter elements to the oncoselectivity of Minute Virus of Mice.Virology. 2007 Apr 25;361(1):174-84. doi: 10.1016/j.virol.2006.11.006. Epub 2006 Dec 18. Virology. 2007. PMID: 17175002 Free PMC article.
References
-
- Astell, C. R., M. Thomson, M. B. Chow, and D. C. Ward. 1983. Structure and replication of minute virus of mice DNA. Cold Spring Harbor Symp. Quant. Biol. 47:751-762. - PubMed
-
- Ball-Goodrich, L. J., R. D. Moir, and P. Tattersall. 1991. Parvoviral target cell specificity: acquisition of fibrotropism by a mutant of the lymphotropic strain of minute virus of mice involves multiple amino acid substitutions within the capsid. Virology 184:175-186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

