PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0
- PMID: 12885887
- PMCID: PMC167235
- DOI: 10.1128/jvi.77.16.8686-8694.2003
PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0
Abstract
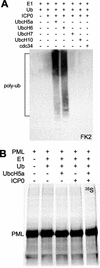
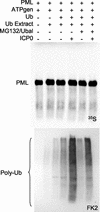
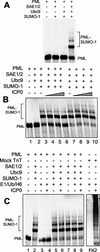
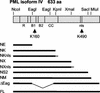
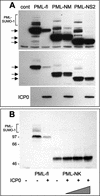
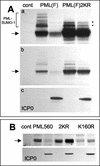
During the early stages of herpes simplex virus type 1 (HSV-1) infection, viral immediate-early regulatory protein ICP0 localizes to and disrupts cellular nuclear structures known as PML nuclear bodies or ND10. These activities correlate with the functions of ICP0 in stimulating lytic infection and reactivating quiescent HSV-1. The disruption of ND10 occurs because ICP0 induces the loss of the SUMO-1-modified forms of PML and the subsequent proteasome-mediated degradation of the PML protein. The functions of ICP0 are largely dependent on the integrity of its zinc-binding RING finger domain. Many RING finger proteins have been found to act as ubiquitin E3 ligase enzymes, stimulating the production of conjugated polyubiquitin chains in the presence of ubiquitin, the ubiquitin-activating enzyme E1, and the appropriate E2 ubiquitin-conjugating enzyme. Substrate proteins that become polyubiquitinated are then subject to degradation by proteasomes. We have previously shown that purified full-length ICP0 acts as an efficient E3 ligase in vitro, producing high-molecular-weight polyubiquitin chains in a RING finger-dependent but substrate-independent manner. In this paper we report on investigations into the factors governing the degradation of PML induced by ICP0 in a variety of in vivo and in vitro assays. We found that ICP0 expression increases the levels of ubiquitinated PML in transfected cells. However, ICP0 does not interact with or directly ubiquitinate either unmodified PML or SUMO-1-modified PML in vitro, suggesting either that additional factors are required for the ICP0-mediated ubiquitination of PML in vivo or that PML degradation is an indirect consequence of some other activity of ICP0 at ND10. Using a transfection-based approach and a family of deletion and point mutations of PML, we found that efficient ICP0-induced PML degradation requires sequences within the C-terminal part of PML and lysine residue 160, one of the principal targets for SUMO-1 modification of the protein.
Figures


References
-
- Ahn, J. H., and G. S. Hayward. 2000. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 274:39-55. - PubMed
-
- Bailey, D., and P. O'Hare. 2002. Herpes simplex virus 1 ICP0 co-localizes with a SUMO-specific protease. J. Gen. Virol. 83:2951-2964. - PubMed
-
- Chelbi-Alix, M. K., and H. de The. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935-941. - PubMed
-
- Duprez, E., A. J. Saurin, J. M. Desterro, V. Lallemand-Breitenbach, K. Howe, M. N. Boddy, E. Solomon, H. de The, R. T. Hay, and P. S. Freemont. 1999. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J. Cell Sci. 112:381-393. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

