De novo synthesis of negative-strand RNA by Dengue virus RNA-dependent RNA polymerase in vitro: nucleotide, primer, and template parameters
- PMID: 12885902
- PMCID: PMC167251
- DOI: 10.1128/jvi.77.16.8831-8842.2003
De novo synthesis of negative-strand RNA by Dengue virus RNA-dependent RNA polymerase in vitro: nucleotide, primer, and template parameters
Erratum in
- J Virol. 2003 Oct;77(19):10730. Padmanbhan R [corrected to Padmanabhan R]
Abstract
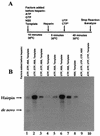
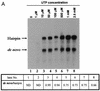
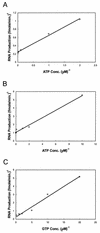
By using a purified dengue virus RNA-dependent RNA polymerase and a subgenomic 770-nucleotide RNA template, it was shown previously that the ratio of the de novo synthesis product to hairpin product formed was inversely proportional to increments of assay temperatures (20 to 40 degrees C). In this study, the components of the de novo preinitiation complex are defined as ATP, a high concentration of GTP (500 micro M), the polymerase, and the template RNA. Even when the 3'-terminal sequence of template RNA was mutated from -GGUUCU-3' to -GGUUUU-3', a high GTP concentration was required for de novo initiation, suggesting that high GTP concentration plays a conformational role. Furthermore, utilization of synthetic primers by the polymerase indicated that AGAA is the optimal primer whereas AG, AGA, and AGAACC were inefficient primers. Moreover, mutational analysis of the highly conserved 3'-terminal dinucleotide CU of the template RNA indicated that change of the 3'-terminal nucleotide from U to C reduced the efficiency about fivefold. The order of preference for the 3'-terminal nucleotide, from highest to lowest, is U, A - G, and C. However, change of the penultimate nucleotide from C to U did not affect the template activity. A model consistent with these results is that the active site of the polymerase switches from a "closed" form, catalyzing de novo initiation through synthesis of short primers, to an "open" form for elongation of a double-stranded template-primer.
Figures



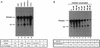


Similar articles
-
Minus-strand initiation by brome mosaic virus replicase within the 3' tRNA-like structure of native and modified RNA templates.J Mol Biol. 1986 Feb 20;187(4):537-46. doi: 10.1016/0022-2836(86)90332-3. J Mol Biol. 1986. PMID: 3754904
-
De novo synthesis of minus strand RNA by the rotavirus RNA polymerase in a cell-free system involves a novel mechanism of initiation.RNA. 2000 Oct;6(10):1455-67. doi: 10.1017/s1355838200001187. RNA. 2000. PMID: 11073221 Free PMC article.
-
De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase.J Biol Chem. 2001 Oct 26;276(43):39926-37. doi: 10.1074/jbc.M104248200. Epub 2001 Aug 23. J Biol Chem. 2001. PMID: 11546770
-
RNA-dependent RNA polymerases from Flaviviridae.Curr Opin Struct Biol. 2009 Dec;19(6):746-51. doi: 10.1016/j.sbi.2009.10.015. Epub 2009 Nov 14. Curr Opin Struct Biol. 2009. PMID: 19914821 Review.
-
Flavivirus RNA synthesis in vitro.Methods. 2015 Dec;91:20-34. doi: 10.1016/j.ymeth.2015.08.002. Epub 2015 Aug 10. Methods. 2015. PMID: 26272247 Free PMC article. Review.
Cited by
-
Molecular basis for nucleotide conservation at the ends of the dengue virus genome.PLoS Pathog. 2012 Sep;8(9):e1002912. doi: 10.1371/journal.ppat.1002912. Epub 2012 Sep 13. PLoS Pathog. 2012. PMID: 23028313 Free PMC article.
-
The first two nucleotides of the respiratory syncytial virus antigenome RNA replication product can be selected independently of the promoter terminus.RNA. 2011 Oct;17(10):1895-906. doi: 10.1261/rna.2813411. Epub 2011 Aug 30. RNA. 2011. PMID: 21878549 Free PMC article.
-
STAT2 signaling and dengue virus infection.JAKSTAT. 2014 Jan 1;3(1):e27715. doi: 10.4161/jkst.27715. Epub 2014 Jan 21. JAKSTAT. 2014. PMID: 24778924 Free PMC article. Review.
-
Dengue virus 2 capsid protein chaperones the strand displacement of 5'-3' cyclization sequences.Nucleic Acids Res. 2021 Jun 4;49(10):5832-5844. doi: 10.1093/nar/gkab379. Nucleic Acids Res. 2021. PMID: 34037793 Free PMC article.
-
Functional RNA elements in the dengue virus genome.Viruses. 2011 Sep;3(9):1739-56. doi: 10.3390/v3091739. Epub 2011 Sep 15. Viruses. 2011. PMID: 21994804 Free PMC article. Review.
References
-
- Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926-39937. - PubMed
-
- Bartelma, G., and R. Padmanabhan. 2002. Expression, purification, and characterization of the RNA 5′-triphosphatase activity of dengue virus type 2 nonstructural protein 3. Virology 299:122-132. - PubMed
-
- Bazan, J. F., and R. J. Fletterick. 1989. Comparative analysis of viral cysteine protease structural models. FEBS Lett. 249:5-7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

