Function of herpes simplex virus type 1 gD mutants with different receptor-binding affinities in virus entry and fusion
- PMID: 12885913
- PMCID: PMC167229
- DOI: 10.1128/jvi.77.16.8962-8972.2003
Function of herpes simplex virus type 1 gD mutants with different receptor-binding affinities in virus entry and fusion
Abstract
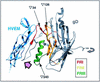
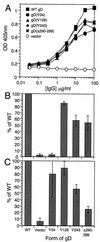
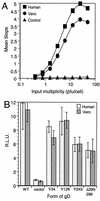
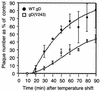
We have studied the receptor-specific function of four linker-insertion mutants of herpes simplex virus type 1 glycoprotein D (gD) representing each of the functional regions of gD. We used biosensor analysis to measure binding of the gD mutants to the receptors HVEM (HveA) and nectin-1 (HveC). One of the mutants, gD(inverted Delta 34t), failed to bind HVEMt but showed essentially wild-type (WT) affinity for nectin-1t. The receptor-binding kinetics and affinities of the other three gD mutants varied over a 1,000-fold range, but each mutant had the same affinity for both receptors. All of the mutants were functionally impaired in virus entry and cell fusion, and the levels of activity were strikingly similar in these two assays. gD(inverted Delta 34)-containing virus was defective on HVEM-expressing cells but did enter nectin-1-expressing cells to about 60% of WT levels. This showed that the defect of this form of gD on HVEM-expressing cells was primarily one of binding and that this was separable from its later function in virus entry. gD(inverted Delta 243t) showed WT binding affinity for both receptors, but virus containing this form of gD had a markedly reduced rate of entry, suggesting that gD(inverted Delta 243) is impaired in a postbinding step in the entry process. There was no correlation between gD mutant activity in fusion or virus entry and receptor-binding affinity. We conclude that gD functions in virus entry and cell fusion regardless of its receptor-binding kinetics and that as long as binding to a functional receptor occurs, entry will progress.
Figures



Similar articles
-
Mutations in the N termini of herpes simplex virus type 1 and 2 gDs alter functional interactions with the entry/fusion receptors HVEM, nectin-2, and 3-O-sulfated heparan sulfate but not with nectin-1.J Virol. 2003 Sep;77(17):9221-31. doi: 10.1128/jvi.77.17.9221-9231.2003. J Virol. 2003. PMID: 12915538 Free PMC article.
-
Structure-based mutagenesis of herpes simplex virus glycoprotein D defines three critical regions at the gD-HveA/HVEM binding interface.J Virol. 2003 Jul;77(14):8127-40. doi: 10.1128/jvi.77.14.8127-8140.2003. J Virol. 2003. PMID: 12829851 Free PMC article.
-
Glycoprotein D homologs in herpes simplex virus type 1, pseudorabies virus, and bovine herpes virus type 1 bind directly to human HveC(nectin-1) with different affinities.Virology. 2001 Feb 1;280(1):7-18. doi: 10.1006/viro.2000.0747. Virology. 2001. PMID: 11162814
-
Different receptors binding to distinct interfaces on herpes simplex virus gD can trigger events leading to cell fusion and viral entry.Virology. 2006 Jan 5;344(1):17-24. doi: 10.1016/j.virol.2005.09.016. Virology. 2006. PMID: 16364731 Review.
-
Viral entry mechanisms: cellular and viral mediators of herpes simplex virus entry.FEBS J. 2009 Dec;276(24):7228-36. doi: 10.1111/j.1742-4658.2009.07402.x. FEBS J. 2009. PMID: 19878306 Free PMC article. Review.
Cited by
-
Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion.Nat Commun. 2011 Dec 6;2:577. doi: 10.1038/ncomms1571. Nat Commun. 2011. PMID: 22146396
-
Receptor Binding-Induced Conformational Changes in Herpes Simplex Virus Glycoprotein D Permit Interaction with the gH/gL Complex to Activate Fusion.Viruses. 2023 Mar 30;15(4):895. doi: 10.3390/v15040895. Viruses. 2023. PMID: 37112875 Free PMC article.
-
The soluble ectodomain of herpes simplex virus gD contains a membrane-proximal pro-fusion domain and suffices to mediate virus entry.Proc Natl Acad Sci U S A. 2004 May 11;101(19):7445-50. doi: 10.1073/pnas.0401883101. Epub 2004 May 3. Proc Natl Acad Sci U S A. 2004. PMID: 15123804 Free PMC article.
-
Multiscale perspectives of virus entry via endocytosis.Virol J. 2013 Jun 5;10:177. doi: 10.1186/1743-422X-10-177. Virol J. 2013. PMID: 23734580 Free PMC article. Review.
-
A herpes simplex virus 2 glycoprotein D mutant generated by bacterial artificial chromosome mutagenesis is severely impaired for infecting neuronal cells and infects only Vero cells expressing exogenous HVEM.J Virol. 2012 Dec;86(23):12891-902. doi: 10.1128/JVI.01055-12. Epub 2012 Sep 19. J Virol. 2012. PMID: 22993162 Free PMC article.
References
-
- Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. - PubMed
-
- Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. - PubMed
-
- Cocchi, F., L. Menotti, P. Mirandola, M. Lopez, and G. Campadelli-Fiume. 1998. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attribute of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 72:9992-10002. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

