Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the noncapsid coding region
- PMID: 12885914
- PMCID: PMC167246
- DOI: 10.1128/jvi.77.16.8973-8984.2003
Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the noncapsid coding region
Abstract
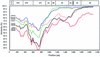
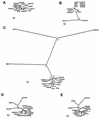
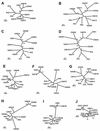
The 65 human enterovirus serotypes are currently classified into five species: Poliovirus (3 serotypes), Human enterovirus A (HEV-A) (12 serotypes), HEV-B (37 serotypes), HEV-C (11 serotypes), and HEV-D (2 serotypes). Coxsackie A virus (CAV) serotypes 1, 11, 13, 15, 17, 18, 19, 20, 21, 22, and 24 constitute HEV-C. We have determined the complete genome sequences for the remaining nine HEV-C serotypes and compared them with the complete sequences of CAV21, CAV24, and the polioviruses. The viruses were most diverse in the capsid region (4 to 36% amino acid difference). A high degree of capsid sequence conservation (96% amino acid identity) suggests that CAV15 and CAV18 should be classified as strains of CAV11 and CAV13, respectively. In the 3CD region, CAV1, CAV19, and CAV22 differed from one another by only 1.2 to 1.4% and CAV11, CAV13, CAV17, CAV20, CAV21, CAV24, and the polioviruses differed from one another by only 1.2 to 3.6%. The two groups, however, differed from one another by 14.6 to 16.2%. The polioviruses as a group were monophyletic only in the capsid region. Only one group of serotypes (CAV1, CAV19, and CAV22) was consistently monophyletic in multiple genome regions. Incongruities among phylogenetic trees based on different genome regions strongly suggest that recombination has occurred between the polioviruses, CAV11, CAV13, CAV17, and CAV20. The close relationship among the polioviruses and CAV11, CAV13, CAV17, CAV20, CAV21, and CAV24 and the uniqueness of CAV1, CAV19, and CAV22 suggest that revisions should be made to the classification of these viruses.
Figures



References
-
- Andersson, P., K. Edman, and A. M. Lindberg. 2002. Molecular analysis of the echovirus 18 prototype. Evidence of interserotypic recombination with echovirus 9. Virus Res. 85:71-83. - PubMed
-
- Cammack, N., A. Phillips, G. Dunn, V. Patel, and P. D. Minor. 1988. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology 167:507-514. - PubMed
-
- Caro, V., S. Guillot, F. Delpeyroux, and R. Crainic. 2001. Molecular strategy for ′serotyping' of human enteroviruses. J. Gen. Virol. 82:79-91. - PubMed
-
- Casas, I., G. F. Palacios, G. Trallero, D. Cisterna, M. C. Freire, and A. Tenorio. 2001. Molecular characterization of human enteroviruses in clinical samples: comparison between VP2, VP1, and RNA polymerase regions using RT nested PCR assays and direct sequencing of products. J. Med. Virol. 65:138-148. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

