Identification of a C-terminal regulatory motif in hepatitis C virus RNA-dependent RNA polymerase: structural and biochemical analysis
- PMID: 12885918
- PMCID: PMC167210
- DOI: 10.1128/jvi.77.16.9020-9028.2003
Identification of a C-terminal regulatory motif in hepatitis C virus RNA-dependent RNA polymerase: structural and biochemical analysis
Abstract
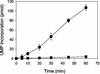
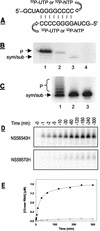
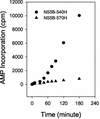
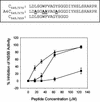
The NS5B RNA-dependent RNA polymerase encoded by the hepatitis C virus (HCV) is a key component of the viral replicase. Reported here is the three-dimensional structure of HCV NS5B polymerase, with the highlight on its C-terminal folding, determined by X-ray crystallography at 2.1-A resolution. Structural analysis revealed that a stretch of C-terminal residues of HCV NS5B inserted into the putative RNA binding cleft, where they formed a hydrophobic pocket and interacted with several important structural elements. This region was found to be conserved and unique to the RNA polymerases encoded by HCV and related viruses. Through biochemical analyses, we confirmed that this region interfered with the binding of HCV NS5B to RNA. Deletion of this fragment from HCV NS5B enhanced the RNA synthesis rate up to approximately 50-fold. These results provide not only direct experimental insights into the role of the C-terminal tail of HCV NS5B polymerase but also a working model for the RNA synthesis mechanism employed by HCV and related viruses.
Figures
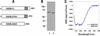




Similar articles
-
Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase.J Virol. 2002 Apr;76(8):3865-72. doi: 10.1128/jvi.76.8.3865-3872.2002. J Virol. 2002. PMID: 11907226 Free PMC article.
-
Evidence for Internal Initiation of RNA Synthesis by the Hepatitis C Virus RNA-Dependent RNA Polymerase NS5B In Cellulo.J Virol. 2019 Sep 12;93(19):e00525-19. doi: 10.1128/JVI.00525-19. Print 2019 Oct 1. J Virol. 2019. PMID: 31315989 Free PMC article.
-
Phosphorylation of hepatitis C virus RNA polymerases ser29 and ser42 by protein kinase C-related kinase 2 regulates viral RNA replication.J Virol. 2014 Oct;88(19):11240-52. doi: 10.1128/JVI.01826-14. Epub 2014 Jul 16. J Virol. 2014. PMID: 25031343 Free PMC article.
-
The discovery of finger loop inhibitors of the hepatitis C virus NS5B polymerase: status and prospects for novel HCV therapeutics.IDrugs. 2006 Jan;9(1):39-43. IDrugs. 2006. PMID: 16374732 Review.
-
Using the Hepatitis C Virus RNA-Dependent RNA Polymerase as a Model to Understand Viral Polymerase Structure, Function and Dynamics.Viruses. 2015 Jul 17;7(7):3974-94. doi: 10.3390/v7072808. Viruses. 2015. PMID: 26193306 Free PMC article. Review.
Cited by
-
Hepatitis C virus RNA replication.Curr Top Microbiol Immunol. 2013;369:167-98. doi: 10.1007/978-3-642-27340-7_7. Curr Top Microbiol Immunol. 2013. PMID: 23463201 Free PMC article. Review.
-
C-Terminal Auto-Regulatory Motif of Hepatitis C Virus NS5B Interacts with Human VAPB-MSP to Form a Dynamic Replication Complex.PLoS One. 2016 Jan 19;11(1):e0147278. doi: 10.1371/journal.pone.0147278. eCollection 2016. PLoS One. 2016. PMID: 26784321 Free PMC article.
-
Assembly, purification, and pre-steady-state kinetic analysis of active RNA-dependent RNA polymerase elongation complex.J Biol Chem. 2012 Mar 23;287(13):10674-10683. doi: 10.1074/jbc.M111.325530. Epub 2012 Feb 2. J Biol Chem. 2012. PMID: 22303022 Free PMC article.
-
RNA polymerase activity and specific RNA structure are required for efficient HCV replication in cultured cells.PLoS Pathog. 2010 Apr 29;6(4):e1000885. doi: 10.1371/journal.ppat.1000885. PLoS Pathog. 2010. PMID: 20442786 Free PMC article.
-
Reconstitution and Functional Analysis of a Full-Length Hepatitis C Virus NS5B Polymerase on a Supported Lipid Bilayer.ACS Cent Sci. 2016 Jul 27;2(7):456-66. doi: 10.1021/acscentsci.6b00112. Epub 2016 Jun 13. ACS Cent Sci. 2016. PMID: 27504492 Free PMC article.
References
-
- Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold. Des. 7:1417-1426. - PubMed
-
- Adachi, T., H. Ago, N. Habuka, K. Okuda, M. Komatsu, S. Ikeda, and K. Yatsunami. 2002. The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase Biochim. Biophys. Acta 1601:38-48. - PubMed
-
- AI, R. H., Y. Xie, Y. Wang, and C. H. Hagedorn. 1998. Expression of recombinant hepatitis C virus non-structural protein 5B in Escherichia coli. Virus Res. 53:141-149. - PubMed
-
- Arnold, J. J., and C. E. Cameron. 2000. Poliovirus RNA-dependent RNA polymerase (3Dpol): assembly of stable, elongation-competent complexes by using a symmetrical primer/template substrate (sym/sub). J. Biol. Chem. 275:5329-5336. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

