Short interfering RNA-mediated interference of gene expression and viral DNA accumulation in cultured plant cells
- PMID: 12886005
- PMCID: PMC170969
- DOI: 10.1073/pnas.1733874100
Short interfering RNA-mediated interference of gene expression and viral DNA accumulation in cultured plant cells
Abstract
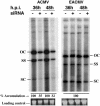
Gene silencing mediated by double-stranded RNA is a sequence-specific RNA degradation mechanism highly conserved in eukaryotes that serves as an antiviral defense pathway in both plants and Drosophila. Short interfering RNAs (siRNAs), the 21- to 23-nt double-stranded intermediates of this natural defense mechanism, are becoming powerful tools for reducing gene expression and countering viral infection in a variety of mammalian cells. Here we report the use of siRNAs to target reporter gene expression and viral DNA accumulation in cultured plant cells. Transient expression of reporter genes encoding either GFP or red fluorescent protein from Discosoma was specifically reduced by 58% and 47%, respectively, at 24 h after codelivery of cognate siRNAs in BY2 protoplasts. In contrast to mammalian systems, the siRNA-induced silencing of GFP expression was transitive as indicated by the presence of siRNAs representing parts of the target RNA outside the region homologous to the triggering siRNA. Codelivery of an siRNA designed to target the mRNA encoding the replication-associated protein (AC1) of the geminivirus African cassava mosaic virus (ACMV) from Cameroon blocked AC1 mRNA accumulation by approximately 91% and inhibited accumulation of the ACMV genomic DNA by approximately 66% at 36 and 48 h after transfection. As with siRNA-induced reporter gene silencing, the siRNA targeting ACMV AC1 was specific and did not affect the replication of East African cassava mosaic Cameroon virus. This report demonstrates the occurrence of siRNA-mediated suppression of gene expression in cultured plant cells and that siRNA can interfere with and suppress accumulation of a nuclear-replicated DNA virus.
Figures



Similar articles
-
Transgenic cassava resistance to African cassava mosaic virus is enhanced by viral DNA-A bidirectional promoter-derived siRNAs.Plant Mol Biol. 2007 Jul;64(5):549-57. doi: 10.1007/s11103-007-9175-6. Epub 2007 May 10. Plant Mol Biol. 2007. PMID: 17492253
-
Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing.J Virol. 2004 Sep;78(17):9487-98. doi: 10.1128/JVI.78.17.9487-9498.2004. J Virol. 2004. PMID: 15308741 Free PMC article.
-
Short interfering RNA accumulation correlates with host recovery in DNA virus-infected hosts, and gene silencing targets specific viral sequences.J Virol. 2004 Jul;78(14):7465-77. doi: 10.1128/JVI.78.14.7465-7477.2004. J Virol. 2004. PMID: 15220420 Free PMC article.
-
Green fluorescent protein specified small interfering RNA-cross-linked iron oxide nanoparticles-Cy5.5.2008 Apr 17 [updated 2008 May 12]. In: Molecular Imaging and Contrast Agent Database (MICAD) [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2004–2013. 2008 Apr 17 [updated 2008 May 12]. In: Molecular Imaging and Contrast Agent Database (MICAD) [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2004–2013. PMID: 20641481 Free Books & Documents. Review.
-
Gene silencing mediated by small interfering RNAs in mammalian cells.Curr Med Chem. 2003 Feb;10(3):245-56. doi: 10.2174/0929867033368493. Curr Med Chem. 2003. PMID: 12570711 Review.
Cited by
-
Enhanced transgene expression in sugarcane by co-expression of virus-encoded RNA silencing suppressors.PLoS One. 2013 Jun 14;8(6):e66046. doi: 10.1371/journal.pone.0066046. Print 2013. PLoS One. 2013. PMID: 23799071 Free PMC article.
-
Multifunctional roles of geminivirus encoded replication initiator protein.Virusdisease. 2019 Mar;30(1):66-73. doi: 10.1007/s13337-018-0458-0. Epub 2018 Jun 9. Virusdisease. 2019. PMID: 31143833 Free PMC article.
-
RNA interference with special reference to combating viruses of crustacea.Indian J Virol. 2012 Sep;23(2):226-43. doi: 10.1007/s13337-012-0084-1. Epub 2012 Aug 14. Indian J Virol. 2012. PMID: 23997446 Free PMC article.
-
Transgenic tobacco plants expressing siRNA targeted against the Mungbean yellow mosaic virus transcriptional activator protein gene efficiently block the viral DNA accumulation.Virusdisease. 2015 Jun;26(1-2):55-61. doi: 10.1007/s13337-015-0251-2. Epub 2015 Apr 18. Virusdisease. 2015. PMID: 26436122 Free PMC article.
-
Emerging threats of begomoviruses to the cultivation of medicinal and aromatic crops and their management strategies.Virusdisease. 2017 Mar;28(1):1-17. doi: 10.1007/s13337-016-0358-0. Epub 2017 Feb 4. Virusdisease. 2017. PMID: 28466050 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

