Transcription increases multiple spontaneous point mutations in Salmonella enterica
- PMID: 12888512
- PMCID: PMC169952
- DOI: 10.1093/nar/gkg651
Transcription increases multiple spontaneous point mutations in Salmonella enterica
Abstract
The spontaneous rate of G.C-->A.T mutations and a hotspot T.A-->G.C transversion are known to increase with the frequency of transcription-increases that have been ascribed primarily to processes that affect only these specific mutations. To investigate how transcription induces other spontaneous point mutations, we tested for its effects in repair-proficient Salmonella enterica using reversion assays of chromosomally inserted alleles. Our results indicate that transcription increases rates of all tested point mutations in the induced gene: induction significantly increased the individual rates of an A.T-->T.A transversion, an A.T-->G.C transition and the pooled rates of the three other point mutations assayed. Although the S.enterica genome is thought to have a mutational bias towards G.C base pairs, transitions creating A.T pairs were approximately 10 times more frequent than the reverse mutation, resulting in an overall mutation pressure to lower G+C contents. Transitions occurred at roughly twice the rate of transversions, similar to results from sequence comparisons; however, several individual transversions are more frequent than the least common transition.
Figures
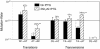
Similar articles
-
Transcribed intergenic regions exhibit a lower frequency of nucleotide polymorphism than the untranscribed intergenic regions in the genomes of Escherichia coli and Salmonella enterica.J Genet. 2023;102:22. J Genet. 2023. PMID: 36823684
-
Detection of mutations in the gyrA gene and class I integron from quinolone-resistant Salmonella enterica serovar Choleraesuis isolates in Taiwan.Vet Microbiol. 2004 Jun 3;100(3-4):247-54. doi: 10.1016/j.vetmic.2004.03.003. Vet Microbiol. 2004. PMID: 15145503
-
Effect of chromosome location on bacterial mutation rates.Mol Biol Evol. 2002 Jan;19(1):85-92. doi: 10.1093/oxfordjournals.molbev.a003986. Mol Biol Evol. 2002. PMID: 11752193
-
Rapid detection of gyrA and parC mutations in quinolone-resistant Salmonella enterica using Pyrosequencing technology.J Microbiol Methods. 2007 Jan;68(1):163-71. doi: 10.1016/j.mimet.2006.07.006. Epub 2006 Aug 24. J Microbiol Methods. 2007. PMID: 16934351
-
Salmonella genomic islands and antibiotic resistance in Salmonella enterica.Future Microbiol. 2010 Oct;5(10):1525-38. doi: 10.2217/fmb.10.122. Future Microbiol. 2010. PMID: 21073312 Review.
Cited by
-
Mutation bias interacts with composition bias to influence adaptive evolution.PLoS Comput Biol. 2020 Sep 28;16(9):e1008296. doi: 10.1371/journal.pcbi.1008296. eCollection 2020 Sep. PLoS Comput Biol. 2020. PMID: 32986712 Free PMC article.
-
Evidence that mutation is universally biased towards AT in bacteria.PLoS Genet. 2010 Sep 9;6(9):e1001115. doi: 10.1371/journal.pgen.1001115. PLoS Genet. 2010. PMID: 20838599 Free PMC article.
-
Spatial Vulnerabilities of the Escherichia coli Genome to Spontaneous Mutations Revealed with Improved Duplex Sequencing.Genetics. 2018 Oct;210(2):547-558. doi: 10.1534/genetics.118.301345. Epub 2018 Aug 3. Genetics. 2018. PMID: 30076202 Free PMC article.
-
Transition bias influences the evolution of antibiotic resistance in Mycobacterium tuberculosis.PLoS Biol. 2019 May 13;17(5):e3000265. doi: 10.1371/journal.pbio.3000265. eCollection 2019 May. PLoS Biol. 2019. PMID: 31083647 Free PMC article.
-
Genetic Variants and Phenotypic Characteristics of Salmonella Typhimurium-Resistant Mutants after Exposure to Carvacrol.Microorganisms. 2020 Jun 22;8(6):937. doi: 10.3390/microorganisms8060937. Microorganisms. 2020. PMID: 32580471 Free PMC article.
References
-
- Brock R.D. (1971) Differential mutation of beta-galactosidase gene of Escherichia coli. Mutat. Res., 11, 181–186. - PubMed
-
- Savic D.J. and Kanazir,D.T. (1972) Effect of a histidine operator constitutive mutation on UV-induced mutability within histidine operon of Salmonella typhimurium. Mol. Gen. Genet., 118, 45–50. - PubMed
-
- Datta A. and Jinks-Robertson,S. (1995) Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science, 268, 1616–1619. - PubMed

