Roles of Saccharomyces cerevisiae DNA polymerases Poleta and Polzeta in response to irradiation by simulated sunlight
- PMID: 12888515
- PMCID: PMC169879
- DOI: 10.1093/nar/gkg489
Roles of Saccharomyces cerevisiae DNA polymerases Poleta and Polzeta in response to irradiation by simulated sunlight
Abstract
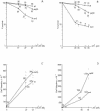
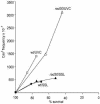
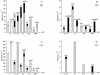
Sunlight causes lesions in DNA that if unrepaired and inaccurately replicated by DNA polymerases yield mutations that result in skin cancer in humans. Two enzymes involved in translesion synthesis (TLS) of UV-induced photolesions are DNA polymerase eta (Poleta) and polymerase zeta (Polzeta), encoded by the RAD30A and REV3 genes, respectively. Previous studies have investigated the TLS roles of these polymerases in human and yeast cells irradiated with monochromatic, short wavelength UVC radiation (254 nm). However, less is known about cellular responses to solar radiation, which is of higher and mixed wavelengths (310-1100 nm) and produces a different spectrum of DNA lesions, including Dewar photoproducts and oxidative lesions. Here we report on the comparative cytotoxic and mutagenic effects of simulated sunlight (SSL) and UVC radiation on yeast wild-type, rad30Delta, rev3Delta and rev3Delta rad30Delta strains. The results with SSL support several previous interpretations on the roles of these two polymerases in TLS of photodimers and (6-4) photoproducts derived from studies with UVC. They further suggest that Poleta participates in the non-mutagenic bypass of SSL-dependent cytosine-containing Dewar photoproducts and 8-oxoguanine, while Polzeta is mainly responsible for the mutagenic bypass of all types of Dewar photoproducts. They also suggest that in the absence of Polzeta, Poleta contributes to UVC- and SSL-induced mutagenesis, possibly by the bypass of photodimers containing deaminated cytosine.
Figures


Similar articles
-
Poleta, Polzeta and Rev1 together are required for G to T transversion mutations induced by the (+)- and (-)-trans-anti-BPDE-N2-dG DNA adducts in yeast cells.Nucleic Acids Res. 2006 Jan 13;34(2):417-25. doi: 10.1093/nar/gkj446. Print 2006. Nucleic Acids Res. 2006. PMID: 16415180 Free PMC article.
-
NGS-based analysis of base-substitution signatures created by yeast DNA polymerase eta and zeta on undamaged and abasic DNA templates in vitro.DNA Repair (Amst). 2017 Nov;59:34-43. doi: 10.1016/j.dnarep.2017.08.011. Epub 2017 Sep 12. DNA Repair (Amst). 2017. PMID: 28946034 Free PMC article.
-
Role of DNA polymerase eta in the bypass of abasic sites in yeast cells.Nucleic Acids Res. 2004 Jul 29;32(13):3984-94. doi: 10.1093/nar/gkh710. Print 2004. Nucleic Acids Res. 2004. PMID: 15284331 Free PMC article.
-
The mechanisms of UV mutagenesis.J Radiat Res. 2011;52(2):115-25. doi: 10.1269/jrr.10175. J Radiat Res. 2011. PMID: 21436607 Review.
-
DNA damage-induced mutation: tolerance via translesion synthesis.Mutat Res. 2000 Jun 30;451(1-2):169-85. doi: 10.1016/s0027-5107(00)00048-8. Mutat Res. 2000. PMID: 10915871 Review.
Cited by
-
DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae.Genetics. 2013 Apr;193(4):1025-64. doi: 10.1534/genetics.112.145219. Genetics. 2013. PMID: 23547164 Free PMC article. Review.
-
A novel variant of DNA polymerase ζ, Rev3ΔC, highlights differential regulation of Pol32 as a subunit of polymerase δ versus ζ in Saccharomyces cerevisiae.DNA Repair (Amst). 2014 Dec;24:138-149. doi: 10.1016/j.dnarep.2014.04.013. Epub 2014 May 10. DNA Repair (Amst). 2014. PMID: 24819597 Free PMC article.
-
The Surprising Diversity of UV-Induced Mutations.Adv Genet (Hoboken). 2024 Mar 7;5(2):2300205. doi: 10.1002/ggn2.202300205. eCollection 2024 Jun. Adv Genet (Hoboken). 2024. PMID: 38884048 Free PMC article.
-
Translesion DNA polymerases Pol zeta, Pol eta, Pol iota, Pol kappa and Rev1 are not essential for repeat-induced point mutation in Neurospora crassa.J Biosci. 2006 Dec;31(5):557-64. doi: 10.1007/BF02708407. J Biosci. 2006. PMID: 17301493
-
Genome-wide maps of UVA and UVB mutagenesis in yeast reveal distinct causative lesions and mutational strand asymmetries.Genetics. 2023 Jul 6;224(3):iyad086. doi: 10.1093/genetics/iyad086. Genetics. 2023. PMID: 37170598 Free PMC article.
References
-
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
-
- Ravanat J.L., Douki,T. and Cadet,J. (2001) Direct and indirect effects of UV radiation on DNA and its components. J. Photochem. Photobiol., 63, 88–102. - PubMed
-
- Sage E. (1999) DNA damage and mutations induced by solar UV radiation. In Baumstark-Khan,C., Kozubek,S. and Horneck,G. (eds), Fundamentals for the Assessment of Risks from Environmental Radiation. Kluwer Academic Publishers, The Netherlands, pp. 115–126.
-
- Brash D.E. (1997) Sunlight and the onset of skin cancer. Trends Genet., 13, 410–414. - PubMed
-
- Stary A. and Sarasin,A. (2002) Molecular mechanisms of UV-induced mutations as revealed by the study of DNA polymerase η in human cells. Res. Microbiol., 153, 441–445. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

