Protein families and TRIBES in genome sequence space
- PMID: 12888524
- PMCID: PMC169885
- DOI: 10.1093/nar/gkg495
Protein families and TRIBES in genome sequence space
Abstract
Accurate detection of protein families allows assignment of protein function and the analysis of functional diversity in complete genomes. Recently, we presented a novel algorithm called TribeMCL for the detection of protein families that is both accurate and efficient. This method allows family analysis to be carried out on a very large scale. Using TribeMCL, we have generated a resource called TRIBES that contains protein family information, comprising annotations, protein sequence alignments and phylogenetic distributions describing 311 257 proteins from 83 completely sequenced genomes. The analysis of at least 60 934 detected protein families reveals that, with the essential families excluded, paralogy levels are similar between prokaryotes, irrespective of genome size. The number of essential families is estimated to be between 366 and 426. We also show that the currently known space of protein families is scale free and discuss the implications of this distribution. In addition, we show that smaller families are often formed by shorter proteins and discuss the reasons for this intriguing pattern. Finally, we analyse the functional diversity of protein families in entire genome sequences. The TRIBES protein family resource is accessible at http://www.ebi.ac.uk/research/cgg/tribes/.
Figures
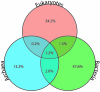



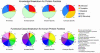
Similar articles
-
On the quality of tree-based protein classification.Bioinformatics. 2005 May 1;21(9):1876-90. doi: 10.1093/bioinformatics/bti244. Epub 2005 Jan 12. Bioinformatics. 2005. PMID: 15647305
-
Automatic annotation of protein function based on family identification.Proteins. 2003 Nov 15;53(3):683-92. doi: 10.1002/prot.10449. Proteins. 2003. PMID: 14579359
-
Identification and distribution of protein families in 120 completed genomes using Gene3D.Proteins. 2005 May 15;59(3):603-15. doi: 10.1002/prot.20409. Proteins. 2005. PMID: 15768405
-
Bioinformatic tools for DNA/protein sequence analysis, functional assignment of genes and protein classification.Appl Microbiol Biotechnol. 2001 Dec;57(5-6):579-92. doi: 10.1007/s00253-001-0844-0. Appl Microbiol Biotechnol. 2001. PMID: 11778865 Review.
-
Towards a covering set of protein family profiles.Prog Biophys Mol Biol. 2000;73(5):321-37. doi: 10.1016/s0079-6107(00)00013-4. Prog Biophys Mol Biol. 2000. PMID: 11063778 Review.
Cited by
-
Comparative genomics of gene-family size in closely related bacteria.Genome Biol. 2004;5(4):R27. doi: 10.1186/gb-2004-5-4-r27. Epub 2004 Mar 18. Genome Biol. 2004. PMID: 15059260 Free PMC article.
-
On the extent and origins of genic novelty in the phylum Nematoda.PLoS Negl Trop Dis. 2008 Jul 2;2(7):e258. doi: 10.1371/journal.pntd.0000258. PLoS Negl Trop Dis. 2008. PMID: 18596977 Free PMC article.
-
Ab Initio Construction and Evolutionary Analysis of Protein-Coding Gene Families with Partially Homologous Relationships: Closely Related Drosophila Genomes as a Case Study.Genome Biol Evol. 2020 Mar 1;12(3):185-202. doi: 10.1093/gbe/evaa041. Genome Biol Evol. 2020. PMID: 32108239 Free PMC article.
-
CGG toolkit: Software components for computational genomics.PLoS Comput Biol. 2023 Nov 7;19(11):e1011498. doi: 10.1371/journal.pcbi.1011498. eCollection 2023 Nov. PLoS Comput Biol. 2023. PMID: 37934729 Free PMC article.
-
The Sulfolobus database.Nucleic Acids Res. 2007 Jan;35(Database issue):D413-5. doi: 10.1093/nar/gkl847. Epub 2006 Nov 6. Nucleic Acids Res. 2007. PMID: 17088281 Free PMC article.
References
-
- Eisenberg D., Marcotte,E.M., Xenarios,I. and Yeates,T.O. (2000) Protein function in the post-genomic era. Nature, 405, 823–826. - PubMed
-
- Tatusov R.L., Koonin,E.V. and Lipman,D.J. (1997) A genomic perspective on protein families. Science, 278, 631–637. - PubMed
-
- Doolittle R.F. (1981) Similar amino acid sequences: chance or common ancestry? Science, 214, 149–159. - PubMed
-
- Devos D. and Valencia,A. (2000) Practical limits of function prediction. Proteins, 41, 98–107. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

