Functional similarity of Knirps CtBP-dependent and CtBP-independent transcriptional repressor activities
- PMID: 12888527
- PMCID: PMC169881
- DOI: 10.1093/nar/gkg491
Functional similarity of Knirps CtBP-dependent and CtBP-independent transcriptional repressor activities
Abstract
Short-range transcriptional repressors are locally acting factors that play important roles in developmental gene expression in Drosophila. To effect repression, Knirps and other short-range repressors bind the CtBP corepressor, but these repressors also function via CtBP-independent pathways. Possible mechanistic differences between CtBP-dependent and -independent repression activities are poorly understood. The distinct activities might provide qualitatively different activities necessary in different promoter contexts, or they might combine to give quantitatively different effects. We analyze separately the CtBP-dependent and CtBP-independent domains of Knirps previously characterized in the embryo to determine possible functional distinctions of the two repression activities. Both domains are active in cell culture and are dependent on the same residues required for activity in the embryo. The domains have similar properties with respect to distance-dependent repression and resistance to inhibition by the deacetylase inhibitor trichostatin A. In tests of repressor-activator specificity, the extent of repression was related not to the chemical nature of the activation domain but to the total activation potential. This result indicates that the balance of competing activation and repression signals is decisive in determining the effectiveness of repressors on genetic switches, suggesting that multiple repression activities are utilized to provide quantitatively, rather than qualitatively, distinct outputs.
Figures


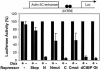

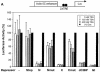

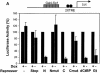




Similar articles
-
CtBP contributes quantitatively to Knirps repression activity in an NAD binding-dependent manner.Mol Cell Biol. 2004 Jul;24(13):5953-66. doi: 10.1128/MCB.24.13.5953-5966.2004. Mol Cell Biol. 2004. PMID: 15199149 Free PMC article.
-
Quantitative contributions of CtBP-dependent and -independent repression activities of Knirps.Development. 2004 May;131(10):2419-29. doi: 10.1242/dev.01075. Development. 2004. PMID: 15128671
-
dCtBP-dependent and -independent repression activities of the Drosophila Knirps protein.Mol Cell Biol. 2000 Oct;20(19):7247-58. doi: 10.1128/MCB.20.19.7247-7258.2000. Mol Cell Biol. 2000. PMID: 10982842 Free PMC article.
-
Transcriptional regulation by C-terminal binding proteins.Int J Biochem Cell Biol. 2007;39(9):1593-607. doi: 10.1016/j.biocel.2007.01.025. Epub 2007 Feb 4. Int J Biochem Cell Biol. 2007. PMID: 17336131 Review.
-
CtBP, an unconventional transcriptional corepressor in development and oncogenesis.Mol Cell. 2002 Feb;9(2):213-24. doi: 10.1016/s1097-2765(02)00443-4. Mol Cell. 2002. PMID: 11864595 Review.
Cited by
-
Functional interaction between the Drosophila knirps short range transcriptional repressor and RPD3 histone deacetylase.J Biol Chem. 2005 Dec 9;280(49):40757-65. doi: 10.1074/jbc.M506819200. Epub 2005 Sep 26. J Biol Chem. 2005. PMID: 16186109 Free PMC article.
-
The Role of CtBP1 in Oncogenic Processes and Its Potential as a Therapeutic Target.Mol Cancer Ther. 2017 Jun;16(6):981-990. doi: 10.1158/1535-7163.MCT-16-0592. Mol Cancer Ther. 2017. PMID: 28576945 Free PMC article. Review.
-
Krüppel-like zinc finger protein Gli-similar 2 (Glis2) represses transcription through interaction with C-terminal binding protein 1 (CtBP1).Nucleic Acids Res. 2005 Dec 2;33(21):6805-15. doi: 10.1093/nar/gki985. Print 2005. Nucleic Acids Res. 2005. PMID: 16326862 Free PMC article.
-
Paradoxical instability-activity relationship defines a novel regulatory pathway for retinoblastoma proteins.Mol Biol Cell. 2010 Nov 15;21(22):3890-901. doi: 10.1091/mbc.E10-06-0520. Epub 2010 Sep 22. Mol Biol Cell. 2010. PMID: 20861300 Free PMC article.
-
Drosophila brakeless interacts with atrophin and is required for tailless-mediated transcriptional repression in early embryos.PLoS Biol. 2007 Jun;5(6):e145. doi: 10.1371/journal.pbio.0050145. PLoS Biol. 2007. PMID: 17503969 Free PMC article.
References
-
- Gray S. and Levine,M. (1996). Transcriptional repression in development. Curr. Opin. Cell Biol., 8, 358–364. - PubMed
-
- Courey A.J. and Jia,S. (2001) Transcriptional repression: the long and the short of it. Genes Dev., 15, 2786–2796. - PubMed
-
- Small S., Arnosti,D.N. and Levine,M. (1993) Spacing ensures autonomous expression of different stripe enhancers in the even-skipped promoter. Development, 119, 767–772. - PubMed
-
- Chen C.K., Kuhnlein,R.P., Eulenberg,K.G., Vincent,S., Affolter,M., and Schuh,R. (1998) The transcription factors KNIRPS and KNIRPS RELATED control cell migration and branch morphogenesis during Drosophila tracheal development. Development, 125, 4959–4968. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

