Parallel competition analysis of Saccharomyces cerevisiae strains differing by a single base using polymerase colonies
- PMID: 12888536
- PMCID: PMC169973
- DOI: 10.1093/nar/gng084
Parallel competition analysis of Saccharomyces cerevisiae strains differing by a single base using polymerase colonies
Abstract
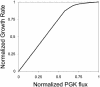
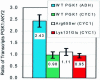
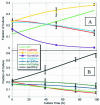
We describe a strategy to analyze the impact of single nucleotide mutations on protein function. Our method utilizes a combination of yeast functional complementation, growth competition of mutant pools and polyacrylamide gel immobilized PCR. A system was constructed in which the yeast PGK1 gene was expressed from a plasmid-borne copy of the gene in a PGK1 deletion strain of Saccharomyces cerevisiae. Using this system, we demonstrated that the enrichment or depletion of PGK1 point mutants from a mixed culture was consistent with the expected results based on the isolated growth rates of the mutants. Enrichment or depletion of individual point mutants was shown to result from increases or decreases, respectively, in the specific activities of the encoded proteins. Further, we demonstrate the ability to analyze the functional effect of many individual point mutations in parallel. By functional complementation of yeast deletions with human homologs, our technique could be readily applied to the functional analysis of single nucleotide polymorphisms in human genes of medical interest.
Figures

References
-
- Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. - PubMed
-
- Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. - PubMed
-
- Masood E. (1999) US firm’s bid to sequence rice genome causes stir in Japan … as consortium plans free SNP map of human genome. Nature, 398, 545–546. - PubMed
-
- Steinmetz L.M., Scharfe,C., Deutschbauer,A.M., Mokranjac,D., Herman,Z.S., Jones,T., Chu,A.M., Giaever,G., Prokisch,H., Oefner,P.J. et al. (2002) Systematic screen for human disease genes in yeast. Nature Genet., 31, 400–404. - PubMed
-
- Winzeler E.A., Shoemaker,D.D., Astromoff,A., Liang,H., Anderson,K., Andre,B., Bangham,R., Benito,R., Boeke,J.D., Bussey,H. et al. (1999) Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

