Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily
- PMID: 12890019
- PMCID: PMC1237129
- DOI: 10.1046/j.1365-2958.2003.03619.x
Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily
Abstract
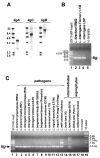
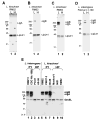
Proteins with bacterial immunoglobulin-like (Big) domains, such as the Yersinia pseudotuberculosis invasin and Escherichia coli intimin, are surface-expressed proteins that mediate host mammalian cell invasion or attachment. Here, we report the identification and characterization of a new family of Big domain proteins, referred to as Lig (leptospiral Ig-like) proteins, in pathogenic Leptospira. Screening of L. interrogans and L. kirschneri expression libraries with sera from leptospirosis patients identified 13 lambda phage clones that encode tandem repeats of the 90 amino acid Big domain. Two lig genes, designated ligA and ligB, and one pseudogene, ligC, were identified. The ligA and ligB genes encode amino-terminal lipoprotein signal peptides followed by 10 or 11 Big domain repeats and, in the case of ligB, a unique carboxy-terminal non-repeat domain. The organization of ligC is similar to that of ligB but contains mutations that disrupt the reading frame. The lig sequences are present in pathogenic but not saprophytic Leptospira species. LigA and LigB are expressed by a variety of virulent leptospiral strains. Loss of Lig protein and RNA transcript expression is correlated with the observed loss of virulence during culture attenuation of pathogenic strains. High-pressure freeze substitution followed by immunocytochemical electron microscopy confirmed that the Lig proteins were localized to the bacterial surface. Immunoblot studies with patient sera found that the Lig proteins are a major antigen recognized during the acute host infection. These observations demonstrate that the Lig proteins are a newly identified surface protein of pathogenic Leptospira, which by analogy to other bacterial immunoglobulin superfamily virulence factors, may play a role in host cell attachment and invasion during leptospiral pathogenesis.
Figures





References
-
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. - PubMed
-
- Ballard SA, Williamson M, Adler B, Vinh T, Faine S. Interactions of virulent and avirulent leptospires with primary cultures of renal epithelial cells. J Med Microbiol. 1986;21:59–67. - PubMed
-
- Ballard SA, Segers RPAM, Bleumink-Pluym N, Fyfe J, Faine S, Adler B. Molecular analysis of the hsp (groE) operon of Leptospira interrogans serovar copenhageni. Mol Microbiol. 1993;8:739–751. - PubMed
-
- Ballard SA, Go M, Segers RP, Adler B. Molecular analysis of the dnaK locus of Leptospira interrogans serovar copenhageni. Gene. 1998;216:21–29. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

