Transcription activation at Escherichia coli FNR-dependent promoters by the gonococcal FNR protein: effects of a novel S18F substitution and comparisons with the corresponding substitution in E. coli FNR
- PMID: 12896992
- PMCID: PMC166479
- DOI: 10.1128/JB.185.16.4734-4747.2003
Transcription activation at Escherichia coli FNR-dependent promoters by the gonococcal FNR protein: effects of a novel S18F substitution and comparisons with the corresponding substitution in E. coli FNR
Abstract
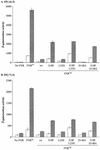
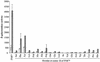
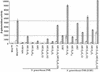
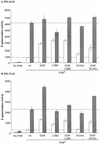
The Neisseria gonorrhoeae genome encodes a homologue of the Escherichia coli FNR protein (the fumarate and nitrate reductase regulator). Despite its similarity to E. coli FNR, the gonococcal FNR only partially complemented an E. coli fnr mutation. After error-prone PCR mutagenesis of the gonococcal fnr gene, we identified four mutant fnr derivatives carrying the same S18F substitution, and we showed that the mutant FNR could activate transcription from a range of class I and class II FNR-dependent promoters in E. coli. Prompted by the similarities between gonococcal and E. coli FNR, we made changes in gonococcal fnr that created substitutions that are equivalent to previously characterized substitutions in E. coli FNR. First, our experiments showed that cysteine, C116, in the gonococcal FNR, equivalent to C122 in E. coli FNR, is essential, presumably because, as in E. coli FNR, it binds to an iron-sulfur center. Second, the L22H and D148A substitutions in gonococcal FNR were made. These changes are equivalent to the L28H and D154A changes in E. coli FNR, which had been shown to increase FNR activity in the presence of oxygen. We show that the effects of these substitutions in gonococcal FNR are distinct from those of the S18F substitution. Similarly, substitutions in the putative activating regions of gonococcal FNR were made. We show that the activity of gonococcal FNR in E. coli can be increased by transplanting certain activating regions from E. coli FNR. The effects of these substitutions are additive to those due to S18F. From these data, we conclude that the effects of the S18F substitution in gonococcal FNR are distinct from the effects of the other substitutions. S18 is immediately adjacent to one of three N-terminal cysteine residues that coordinate the iron-sulfur center, and thus the S18F substitution is most likely to stabilize this center. Support for this came from complementary experiments in which we created the S24F substitution in E. coli FNR, which is equivalent to the S18F substitution in gonococcal FNR. Our results show that the S24F substitution changes the activity of E. coli FNR and that the changes are distinct from those due to previously characterized substitutions.
Figures




References
-
- Bates, D. M., C. V. Popescu, N. Khoroshilova, K. Vogt, H. Beinert, E. Münck, and P. J. Kiley. 2000. Substitution of leucine 28 with a histidine in the Escherichia coli transcription factor FNR results in increased stability of the [4Fe-4S]2+ cluster to oxygen. J. Biol. Chem. 275:6234-6240. - PubMed
-
- Browning, D. F., C. M. Beatty, A. J. Wolfe, J. A. Cole, and S. J. Busby. 2002. Independent regulation of the divergent Escherichia coli nrfA and acsP1 promoters by a nucleoprotein assembly at a shared regulatory region. Mol. Microbiol. 43:687-701. - PubMed
-
- Browning, D. F., D. Lee, J. Green, and S. J. Busby. 2002. Secrets of bacterial transcription initiation taught by the Escherichia coli FNR protein, p. 127-142. In D. A. Hodgson and C. M. Thomas (ed.), Signals, switches, regulons, and cascades: control of bacterial gene expression. Proceedings of the 150th ordinary meeting. Society for General Microbiology, Reading, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

