Epigenetic properties and identification of an imprint mark in the Nesp-Gnasxl domain of the mouse Gnas imprinted locus
- PMID: 12897124
- PMCID: PMC166348
- DOI: 10.1128/MCB.23.16.5475-5488.2003
Epigenetic properties and identification of an imprint mark in the Nesp-Gnasxl domain of the mouse Gnas imprinted locus
Abstract
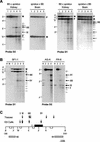
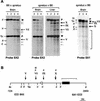
The Gnas locus in the mouse is imprinted with a complex arrangement of alternative transcripts defined by promoters with different patterns of monoallelic expression. The Gnas transcript is subject to tissue-specific imprinted expression, Nesp is expressed only from the maternal allele, and Gnasxl is expressed only from the paternal allele. The mechanisms controlling these expression patterns are not known. To identify potential imprinting regulatory regions, particularly for the reciprocally expressed Nesp and Gnasxl promoters, we examined epigenetic properties of the locus in gametes, embryonic stem cells, and fetal and adult tissues. The Nesp and Gnasxl promoter regions are contained in extensive CpG islands with methylation of the paternal allele at Nesp and the maternal allele at Gnasxl. Parental allele-specific DNase I-hypersensitive sites were found at these regions, which correlate with hypomethylation rather than actual expression status. A germ line methylation mark was identified covering the promoters for Gnasxl and the antisense transcript Nespas. Prominent DNase I-hypersensitive sites present on paternal alleles in embryonic stem cells are contained within this mark. This is the second gametic mark identified at Gnas and suggests that the Nesp and Gnasxl promoters are under separate control from the Gnas promoter. We propose models to account for the regulation of imprinting at the locus.
Figures





References
-
- Arnaud, P., D. Monk, M. Hitchins, E. Gordon, W. Dean, C. V. Beechey, J. Peters, W. Craigen, M. Preece, P. Stanier, G. E. Moore, G. E., and G. Kelsey. 2003. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum. Mol. Genet. 12:1005-1019. - PubMed
-
- Ball, S. T., C. M. Williamson, C. Hayes, T. Hacker, and J. Peters. 2001. The spatial and temporal expression pattern of Nesp and its antisense Nespas, in mid-gestation mouse embryos. Mech. Dev. 100:79-81. - PubMed
-
- Bastepe, M., J. E. Pincus, T. Sugimoto, K. Tojo, M. Kanatani, Y. Azuma, K. Kruse, A. L. Rosenbloom, H. Koshiyama, and H. Jüppner. 2001. Positional dissociation between the genetic mutation responsible for pseudohypoparathyroidism type 1b and the associated methylation defect at exon A/B: evidence for a long-range regulatory element within the imprinted GNAS1 locus. Hum. Mol. Genet. 10:1231-1241. - PubMed
-
- Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
