Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae
- PMID: 12897126
- PMCID: PMC166345
- DOI: 10.1128/MCB.23.16.5502-5515.2003
Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae
Abstract
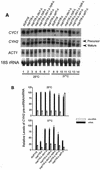
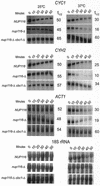
A nuclear mRNA degradation (DRN) system was identified from analysis of mRNA turnover rates in nup116-Delta strains of Saccharomyces cerevisiae lacking the ability to export all RNAs, including poly(A) mRNAs, at the restrictive temperature. Northern blotting, in situ hybridization, and blocking transcription with thiolutin in nup116-delta strains revealed a rapid degradation of mRNAs in the nucleus that was suppressed by the rrp6-delta, rai1-delta, and cbc1-delta deletions, but not by the upf1-delta deletion, suggesting that DRN requires Rrp6p, a 3'-to-5' nuclear exonuclease, the Rat1p, a 5'-to-3' nuclear exonuclease, and Cbc1p, a component of CBC, the nuclear cap binding complex, which may direct the mRNAs to the site of degradation. We propose that certain normal mRNAs retained in the nucleus are degraded by the DRN system, similar to degradation of transcripts with 3' end formation defects in certain mutants.
Figures






References
-
- Bailer, S. M., C. Balduf, J. Katahira, A. Podtelejnikov, C. Rollenhagen, M. Mann, N. Pante, and E. Hurt. 2000. Nup116p associates with the Nup82p-Nsp1p-Nup159p nucleoporin complex. J. Biol. Chem. 275:23540-23548. - PubMed
-
- Beelman, C. A., A. Stevens, G. Caponigro, T. E. LaGrandeur, L. Hatfield, D. M. Fortner, and R. Parker. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382:642-646. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
