Stem-loop IV of tetrahymena telomerase RNA stimulates processivity in trans
- PMID: 12897134
- PMCID: PMC166324
- DOI: 10.1128/MCB.23.16.5606-5613.2003
Stem-loop IV of tetrahymena telomerase RNA stimulates processivity in trans
Abstract
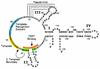
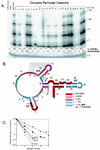
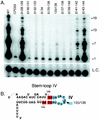
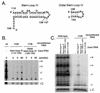
Telomerase is a ribonucleoprotein enzyme responsible for the addition of telomeres onto the ends of chromosomes. Short or dysfunctional telomeres can lead to cell growth arrest, apoptosis, and genomic instability. Telomerase uses its RNA subunit to copy a short template region for telomere synthesis. To probe for regions of Tetrahymena telomerase RNA essential for function, we assayed 27 circularly permuted RNA deletions for telomerase in vitro activity and binding to the telomerase reverse transcriptase catalytic protein subunit. We found that stem-loop IV is required for wild-type telomerase activity in vitro and will stimulate processivity when added in trans.
Figures

Similar articles
-
Roles for RNA in telomerase nucleotide and repeat addition processivity.Mol Cell. 2003 Jun;11(6):1673-83. doi: 10.1016/s1097-2765(03)00232-6. Mol Cell. 2003. PMID: 12820978 Free PMC article.
-
Structure of stem-loop IV of Tetrahymena telomerase RNA.EMBO J. 2006 Jul 12;25(13):3156-66. doi: 10.1038/sj.emboj.7601195. Epub 2006 Jun 15. EMBO J. 2006. PMID: 16778765 Free PMC article.
-
Template boundary definition in Tetrahymena telomerase.Genes Dev. 2002 Feb 15;16(4):415-20. doi: 10.1101/gad.962602. Genes Dev. 2002. PMID: 11850404 Free PMC article.
-
Structure and function of telomerase RNA.Curr Opin Struct Biol. 2006 Jun;16(3):307-18. doi: 10.1016/j.sbi.2006.05.005. Epub 2006 May 18. Curr Opin Struct Biol. 2006. PMID: 16713250 Review.
-
Telomerase Mechanism of Telomere Synthesis.Annu Rev Biochem. 2017 Jun 20;86:439-460. doi: 10.1146/annurev-biochem-061516-045019. Epub 2017 Jan 30. Annu Rev Biochem. 2017. PMID: 28141967 Free PMC article. Review.
Cited by
-
Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function.RNA. 2006 Aug;12(8):1475-85. doi: 10.1261/rna.112306. Epub 2006 Jun 29. RNA. 2006. PMID: 16809815 Free PMC article.
-
The structure of an enzyme-activating fragment of human telomerase RNA.RNA. 2005 Apr;11(4):394-403. doi: 10.1261/rna.7222505. Epub 2005 Feb 9. RNA. 2005. PMID: 15703438 Free PMC article.
-
Characterisation of the Arabidopsis thaliana telomerase TERT-TR complex.Plant Mol Biol. 2024 May 14;114(3):56. doi: 10.1007/s11103-024-01461-w. Plant Mol Biol. 2024. PMID: 38743198 Free PMC article.
-
The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis.Nat Struct Mol Biol. 2011 Nov 20;18(12):1371-5. doi: 10.1038/nsmb.2174. Nat Struct Mol Biol. 2011. PMID: 22101935 Free PMC article.
-
Telomerase: an RNP enzyme synthesizes DNA.Cold Spring Harb Perspect Biol. 2011 May 1;3(5):a003558. doi: 10.1101/cshperspect.a003558. Cold Spring Harb Perspect Biol. 2011. PMID: 20660025 Free PMC article. Review.
References
-
- Autexier, C., and C. W. Greider. 1995. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev. 9:2227-2239. - PubMed
-
- Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 1998. Reconstitution of human telomerase activity in vitro. Curr. Biol. 8:177-180. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
