Mouse mammary tumor virus c-rel transgenic mice develop mammary tumors
- PMID: 12897145
- PMCID: PMC166341
- DOI: 10.1128/MCB.23.16.5738-5754.2003
Mouse mammary tumor virus c-rel transgenic mice develop mammary tumors
Abstract
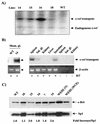
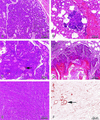
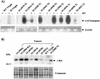
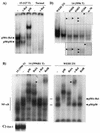
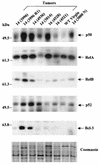
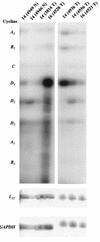
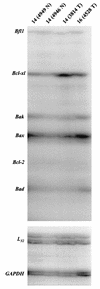
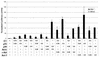
Amplification, overexpression, or rearrangement of the c-rel gene, encoding the c-Rel NF-kappaB subunit, has been reported in solid and hematopoietic malignancies. For example, many primary human breast cancer tissue samples express high levels of nuclear c-Rel. While the Rev-T oncogene v-rel causes tumors in birds, the ability of c-Rel to transform in vivo has not been demonstrated. To directly test the role of c-Rel in breast tumorigenesis, mice were generated in which overexpression of mouse c-rel cDNA was driven by the hormone-responsive mouse mammary tumor virus long terminal repeat (MMTV-LTR) promoter, and four founder lines identified. In the first cycle of pregnancy, the expression of transgenic c-rel mRNA was observed, and levels of c-Rel protein were increased in the mammary gland. Importantly, 31.6% of mice developed one or more mammary tumors at an average age of 19.9 months. Mammary tumors were of diverse histology and expressed increased levels of nuclear NF-kappaB. Analysis of the composition of NF-kappaB complexes in the tumors revealed aberrant nuclear expression of multiple subunits, including c-Rel, p50, p52, RelA, RelB, and the Bcl-3 protein, as observed previously in human primary breast cancers. Expression of the cancer-related NF-kappaB target genes cyclin D1, c-myc, and bcl-xl was significantly increased in grossly normal transgenic mammary glands starting the first cycle of pregnancy and increased further in mammary carcinomas compared to mammary glands from wild-type mice or virgin transgenic mice. In transient transfection analysis in untransformed breast epithelial cells, c-Rel-p52 or -p50 heterodimers either potently or modestly induced cyclin D1 promoter activity, respectively. Lastly, stable overexpression of c-Rel resulted in increased cyclin D1 and NF-kappaB p52 and p50 subunit protein levels. These results indicate for the first time that dysregulated expression of c-Rel, as observed in breast cancers, is capable of contributing to mammary tumorigenesis.
Figures




Similar articles
-
RelB/p52 NF-kappaB complexes rescue an early delay in mammary gland development in transgenic mice with targeted superrepressor IkappaB-alpha expression and promote carcinogenesis of the mammary gland.Mol Cell Biol. 2005 Nov;25(22):10136-47. doi: 10.1128/MCB.25.22.10136-10147.2005. Mol Cell Biol. 2005. PMID: 16260626 Free PMC article.
-
7,12-dimethylbenz(a)anthracene treatment of a c-rel mouse mammary tumor cell line induces epithelial to mesenchymal transition via activation of nuclear factor-kappaB.Cancer Res. 2006 Mar 1;66(5):2570-5. doi: 10.1158/0008-5472.CAN-05-3056. Cancer Res. 2006. PMID: 16510574
-
Inducible IkappaB kinase/IkappaB kinase epsilon expression is induced by CK2 and promotes aberrant nuclear factor-kappaB activation in breast cancer cells.Cancer Res. 2005 Dec 15;65(24):11375-83. doi: 10.1158/0008-5472.CAN-05-1602. Cancer Res. 2005. PMID: 16357145
-
MMTV mouse models and the diagnostic values of MMTV-like sequences in human breast cancer.Expert Rev Mol Diagn. 2009 Jul;9(5):423-40. doi: 10.1586/erm.09.31. Expert Rev Mol Diagn. 2009. PMID: 19580428 Free PMC article. Review.
-
NF-kappaB and cell-cycle regulation: the cyclin connection.Cytokine Growth Factor Rev. 2001 Mar;12(1):73-90. doi: 10.1016/s1359-6101(00)00018-6. Cytokine Growth Factor Rev. 2001. PMID: 11312120 Review.
Cited by
-
Gene expression profiling in sinonasal adenocarcinoma.BMC Med Genomics. 2009 Nov 10;2:65. doi: 10.1186/1755-8794-2-65. BMC Med Genomics. 2009. PMID: 19903339 Free PMC article.
-
Elevated EDAR signalling promotes mammary gland tumourigenesis with squamous metaplasia.Oncogene. 2022 Feb;41(7):1040-1049. doi: 10.1038/s41388-021-01902-6. Epub 2021 Dec 16. Oncogene. 2022. PMID: 34916592 Free PMC article.
-
Survival and differentiation of mammary epithelial cells in mammary gland development require nuclear retention of Id2 due to RANK signaling.Mol Cell Biol. 2011 Dec;31(23):4775-88. doi: 10.1128/MCB.05646-11. Epub 2011 Sep 26. Mol Cell Biol. 2011. PMID: 21947283 Free PMC article.
-
NFkappaB1/p50 is not required for tumor necrosis factor-stimulated growth of primary mammary epithelial cells: implications for NFkappaB2/p52 and RelB.Endocrinology. 2007 Jan;148(1):268-78. doi: 10.1210/en.2006-0500. Epub 2006 Sep 28. Endocrinology. 2007. PMID: 17008396 Free PMC article.
-
RelB/p52 NF-kappaB complexes rescue an early delay in mammary gland development in transgenic mice with targeted superrepressor IkappaB-alpha expression and promote carcinogenesis of the mammary gland.Mol Cell Biol. 2005 Nov;25(22):10136-47. doi: 10.1128/MCB.25.22.10136-10147.2005. Mol Cell Biol. 2005. PMID: 16260626 Free PMC article.
References
-
- Abbadie, C., N. Kabrun, F. Bouali, J. Smardova, D. Stehelin, B. Vandenbunder, and P. J. Enrietto. 1993. High levels of c-rel expression are associated with programmed cell death in the developing avian embryo and in bone marrow cells in vitro. Cell 75:899-912. - PubMed
-
- Alizadeh, A. A., M. B. Eisen, R. E. Davis, C. Ma, I. S. Lossos, A. Rosenwald, J. C. Boldrick, H. Sabet, T. Tran, X. Yu, J. I. Powell, L. Yang, G. E. Marti, T. Moore, J. Hudson, Jr., L. Lu, D. B. Lewis, R. Tibshirani, G. Sherlock, W. C. Chan, T. C. Greiner, D. D. Weisenburger, J. O. Armitage, R. Warnke, L. M. Staudt, and et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403:503-511. - PubMed
-
- Arsura, M., M. J. FitzGerald, N. Fausto, and G. E. Sonenshein. 1997. Nuclear factor-κB/Rel blocks transforming growth factor beta1-induced apoptosis of murine hepatocyte cell lines. Cell Growth Differ. 8:1049-1059. - PubMed
-
- Arsura, M., and G. E. Sonenshein. 2001. The roles of the c-myc and c-myb oncogenes in hematopoiesis and leukemogenesis, p. 521-549. In K. Ravid and J. Licht (ed.), Transcription factors: normal and malignant development of blood cells. Wiley-Liss Inc., New York, N.Y.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
