Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity
- PMID: 12897151
- PMCID: PMC166325
- DOI: 10.1128/MCB.23.16.5816-5824.2003
Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity
Abstract
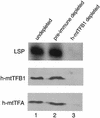
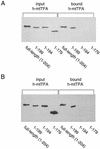
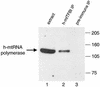
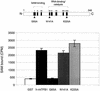
A significant advancement in understanding mitochondrial gene expression is the recent identification of two new human mitochondrial transcription factors, h-mtTFB1 and h-mtTFB2. Both proteins stimulate transcription in collaboration with the high-mobility group box transcription factor, h-mtTFA, and are homologous to rRNA methyltransferases. In fact, the dual-function nature of h-mtTFB1 was recently demonstrated by its ability to methylate a conserved rRNA substrate. Here, we demonstrate that h-mtTFB1 binds h-mtTFA both in HeLa cell mitochondrial extracts and in direct-binding assays via an interaction that requires the C-terminal tail of h-mtTFA, a region necessary for transcriptional activation. In addition, point mutations in conserved methyltransferase motifs of h-mtTFB1 revealed that it stimulates transcription in vitro independently of S-adenosylmethionine binding and rRNA methyltransferase activity. Furthermore, one mutation (G65A) eliminated the ability of h-mtTFB1 to bind DNA yet did not affect transcriptional activation. These results, coupled with the observation that h-mtTFB1 and human mitochondrial RNA (h-mtRNA) polymerase can also be coimmunoprecipitated, lead us to propose a model in which h-mtTFA demarcates mitochondrial promoter locations and where h-mtTFB proteins bridge an interaction between the C-terminal tail of h-mtTFA and mtRNA polymerase to facilitate specific initiation of transcription. Altogether, these data provide important new insight into the mechanism of transcription initiation in human mitochondria and indicate that the dual functions of h-mtTFB1 can be separated.
Figures

References
-
- Bogenhagen, D. F. 1996. Interaction of mtTFB and mtRNA polymerase at core promoters for transcription of Xenopus laevis mtDNA. J. Biol. Chem. 271:12036-12041. - PubMed
-
- Bussiere, D. E., S. W. Muchmore, C. G. Dealwis, G. Schluckebier, V. L. Nienaber, R. P. Edalji, K. A. Walter, U. S. Ladror, T. F. Holzman, and C. Abad-Zapatero. 1998. Crystal structure of ErmC′, an rRNA methyltransferase which mediates antibiotic resistance in bacteria. Biochemistry 37:7103-7112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
