The mitochondrial genome of Chara vulgaris: insights into the mitochondrial DNA architecture of the last common ancestor of green algae and land plants
- PMID: 12897260
- PMCID: PMC167177
- DOI: 10.1105/tpc.013169
The mitochondrial genome of Chara vulgaris: insights into the mitochondrial DNA architecture of the last common ancestor of green algae and land plants
Abstract
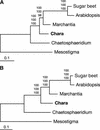
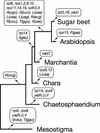
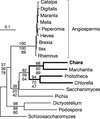
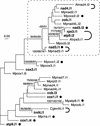
Mitochondrial DNA (mtDNA) has undergone radical changes during the evolution of green plants, yet little is known about the dynamics of mtDNA evolution in this phylum. Land plant mtDNAs differ from the few green algal mtDNAs that have been analyzed to date by their expanded size, long spacers, and diversity of introns. We have determined the mtDNA sequence of Chara vulgaris (Charophyceae), a green alga belonging to the charophycean order (Charales) that is thought to be the most closely related alga to land plants. This 67,737-bp mtDNA sequence, displaying 68 conserved genes and 27 introns, was compared with those of three angiosperms, the bryophyte Marchantia polymorpha, the charophycean alga Chaetosphaeridium globosum (Coleochaetales), and the green alga Mesostigma viride. Despite important differences in size and intron composition, Chara mtDNA strikingly resembles Marchantia mtDNA; for instance, all except 9 of 68 conserved genes lie within blocks of colinear sequences. Overall, our genome comparisons and phylogenetic analyses provide unequivocal support for a sister-group relationship between the Charales and the land plants. Only four introns in land plant mtDNAs appear to have been inherited vertically from a charalean algar ancestor. We infer that the common ancestor of green algae and land plants harbored a tightly packed, gene-rich, and relatively intron-poor mitochondrial genome. The group II introns in this ancestral genome appear to have spread to new mtDNA sites during the evolution of bryophytes and charalean green algae, accounting for part of the intron diversity found in Chara and land plant mitochondria.
Figures



References
-
- Adachi, J., and Hasegawa, M. (1996). MOLPHY version 2.3: Programs for molecular phylogenetics based on maximum likelihood method. Comput. Sci. Monogr. 28, 1–150.
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. - PubMed
-
- Beckert, S., Muhle, H., Pruchner, D., and Knoop, V. (2001). The mitochondrial nad2 gene as a novel marker locus for phylogenetic analysis of early land plants: A comparative analysis in mosses. Mol. Phylogenet. Evol. 18, 117–126. - PubMed
-
- Beckert, S., Steinhauser, S., Muhle, H., and Knoop, V. (1999). A molecular phylogeny of bryophytes based on nucleotide sequences of the mitochondrial nad5 gene. Plant Syst. Evol. 218, 179–192.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

