Rapid identification of Candida species by using nuclear magnetic resonance spectroscopy and a statistical classification strategy
- PMID: 12902244
- PMCID: PMC169103
- DOI: 10.1128/AEM.69.8.4566-4574.2003
Rapid identification of Candida species by using nuclear magnetic resonance spectroscopy and a statistical classification strategy
Abstract
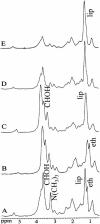
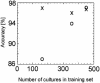
Nuclear magnetic resonance (NMR) spectra were acquired from suspensions of clinically important yeast species of the genus Candida to characterize the relationship between metabolite profiles and species identification. Major metabolites were identified by using two-dimensional correlation NMR spectroscopy. One-dimensional proton NMR spectra were analyzed by using a staged statistical classification strategy. Analysis of NMR spectra from 442 isolates of Candida albicans, C. glabrata, C. krusei, C. parapsilosis, and C. tropicalis resulted in rapid, accurate identification when compared with conventional and DNA-based identification. Spectral regions used for the classification of the five yeast species revealed species-specific differences in relative amounts of lipids, trehalose, polyols, and other metabolites. Isolates of C. parapsilosis and C. glabrata with unusual PCR fingerprinting patterns also generated atypical NMR spectra, suggesting the possibility of intraspecies discontinuity. We conclude that NMR spectroscopy combined with a statistical classification strategy is a rapid, nondestructive, and potentially valuable method for identification and chemotaxonomic characterization that may be broadly applicable to fungi and other microorganisms.
Figures
References
-
- Botha, A., and J. L. F. Kock. 1993. Application of fatty acid profiles in the identification of yeasts. Int. J. Food Microbiol. 19:39-51. - PubMed
-
- Bubb, W. A., L. C. Wright, M. Cagney, R. T. Santangelo, T. C. Sorrell, and P. W. Kuchel. 1999. Heteronuclear NMR studies of metabolites produced by Cryptococcus neoformans in culture media: identification of possible virulence factors. Magn. Reson. Med. 42:442-453. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

