Tropheryma whipplei Twist: a human pathogenic Actinobacteria with a reduced genome
- PMID: 12902375
- PMCID: PMC403771
- DOI: 10.1101/gr.1474603
Tropheryma whipplei Twist: a human pathogenic Actinobacteria with a reduced genome
Abstract
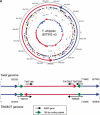
The human pathogen Tropheryma whipplei is the only known reduced genome species (<1 Mb) within the Actinobacteria [high G+C Gram-positive bacteria]. We present the sequence of the 927303-bp circular genome of T. whipplei Twist strain, encoding 808 predicted protein-coding genes. Specific genome features include deficiencies in amino acid metabolisms, the lack of clear thioredoxin and thioredoxin reductase homologs, and a mutation in DNA gyrase predicting a resistance to quinolone antibiotics. Moreover, the alignment of the two available T. whipplei genome sequences (Twist vs. TW08/27) revealed a large chromosomal inversion the extremities of which are located within two paralogous genes. These genes belong to a large cell-surface protein family defined by the presence of a common repeat highly conserved at the nucleotide level. The repeats appear to trigger frequent genome rearrangements in T. whipplei, potentially resulting in the expression of different subsets of cell surface proteins. This might represent a new mechanism for evading host defenses. The T. whipplei genome sequence was also compared to other reduced bacterial genomes to examine the generality of previously detected features. The analysis of the genome sequence of this previously largely unknown human pathogen is now guiding the development of molecular diagnostic tools and more convenient culture conditions.
Figures





References
-
- Akman, L., Yamashita, A., Watanabe, H., Oshima, K., Shiba, T., Hattori, M., and Aksoy, S. 2002. Genome sequence of the endocellular obligate symbiot of tsetse flies, Wiggleworthia glossinidia. Nat. Genet. 32: 402-407. - PubMed
-
- Andersson, J.O. and Andersson, S.G.E. 2001. Pseudogenes, junk DNA, and the dynamics of Rickettsia genomes. Mol. Biol. Evol. 18: 829-839. - PubMed
-
- Andersson, S.G., Zomorodipour, A., Andersson, J.O., Sicheritz-Ponten, T., Alsmark, U.C., Podowski, R.M., Naslund, A.K., Eriksson, A.S., Winkler, H.H., and Kurland, C.G. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396: 133-140. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
