Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes
- PMID: 12902380
- PMCID: PMC403777
- DOI: 10.1101/gr.1272403
Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes
Abstract
Although mRNA decay rates are a key determinant of the steady-state concentration for any given mRNA species, relatively little is known, on a population level, about what factors influence turnover rates and how these rates are integrated into cellular decisions. We decided to measure mRNA decay rates in two human cell lines with high-density oligonucleotide arrays that enable the measurement of decay rates simultaneously for thousands of mRNA species. Using existing annotation and the Gene Ontology hierarchy of biological processes, we assign mRNAs to functional classes at various levels of resolution and compare the decay rate statistics between these classes. The results show statistically significant organizational principles in the variation of decay rates among functional classes. In particular, transcription factor mRNAs have increased average decay rates compared with other transcripts and are enriched in "fast-decaying" mRNAs with half-lives <2 h. In contrast, we find that mRNAs for biosynthetic proteins have decreased average decay rates and are deficient in fast-decaying mRNAs. Our analysis of data from a previously published study of Saccharomyces cerevisiae mRNA decay shows the same functional organization of decay rates, implying that it is a general organizational scheme for eukaryotes. Additionally, we investigated the dependence of decay rates on sequence composition, that is, the presence or absence of short mRNA motifs in various regions of the mRNA transcript. Our analysis recovers the positive correlation of mRNA decay with known AU-rich mRNA motifs, but we also uncover further short mRNA motifs that show statistically significant correlation with decay. However, we also note that none of these motifs are strong predictors of mRNA decay rate, indicating that the regulation of mRNA decay is more complex and may involve the cooperative binding of several RNA-binding proteins at different sites.
Figures



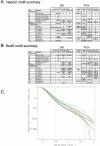
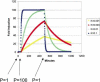
References
-
- Chen, C.Y., Gherzi, R., Ong, S.E., Chan, E.L., Raijmakers, R., Pruijn, G.J., Stoecklin, G., Moroni, C., Mann, M., and Karin, M. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451-464. - PubMed
-
- Darnell Jr., J.E. 1982. Variety in the level of gene control in eukaryotic cells. Nature 297: 365-371. - PubMed
WEB SITE REFERENCES
-
- http://genomes.rockefeller.edu/~yange/; decay rate estimates for 5245 accessions.
-
- http://genome-www.stanford.edu/Saccharomyces; Saccharomyces cerevisiae database.
-
- http://genome-www.stanford.edu/turnover; turnover cDNA microarray data from Saccharomyces cerevisiae.
-
- http://www.affymetrix.com/analysis/index.affx; Affymetrix.
-
- http://www.geneontology.org; Gene Ontology Consortium.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
