Detection of clarithromycin-resistant Helicobacter pylori in stool samples
- PMID: 12904368
- PMCID: PMC179782
- DOI: 10.1128/JCM.41.8.3636-3640.2003
Detection of clarithromycin-resistant Helicobacter pylori in stool samples
Abstract
The recognition of the role of Helicobacter pylori in gastric diseases has led to the widespread use of antibiotics in the eradication of this pathogen. The most advocated therapy, triple therapy, often includes clarithromycin. It is well known that clarithromycin resistance is one of the major causes of eradication failure. The development of a rapid noninvasive technique that could easily be performed on fecal samples and that could also provide information about the antibiotic resistance of this microorganism is therefore advisable. Previous findings have demonstrated that clarithromycin resistance is due to a single point mutation in the 23S rRNA. All the mutations described have been associated with specific restriction sites, namely BsaI (A2143G), MboII (A2142C/G), and HhaI (T2717C). On this basis we have developed a new method, a seminested PCR, allowing screening for clarithromycin resistance of H. pylori directly on stool samples. This method furnished a 783-bp fragment of the 23S rRNA, which was subsequently digested by MboII, BsaI, and HhaI, in order to identify single point mutations associated with clarithromycin resistance. Of a total of 283 stool samples examined, 125 were H. pylori positive and two of them were shown to contain clarithromycin-resistant strains due to the presence of a mutation at position 2717, whereas no PCR products contained mutations at position 2142 or 2143. In order to evaluate the reliability of the new system, we compared the results of restriction analysis of the PCR products with the MICs shown by the H. pylori isolates by culturing gastric biopsies from the same patients.
Figures
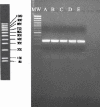
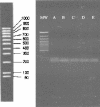

Similar articles
-
Polymerase chain reaction--restriction fragment length polymorphism analysis of clarithromycin-resistant Helicobacter pylori infection in children using stool sample.Helicobacter. 2005 Jun;10(3):205-13. doi: 10.1111/j.1523-5378.2005.00312.x. Helicobacter. 2005. PMID: 15904478
-
Genotyping and antimicrobial resistance patterns of Helicobacter pylori in human and dogs associated with A2142G and A2143G point mutations in clarithromycin resistance.Microb Pathog. 2018 Oct;123:330-338. doi: 10.1016/j.micpath.2018.07.016. Epub 2018 Jul 18. Microb Pathog. 2018. PMID: 30031039
-
Low Helicobacter pylori primary resistance to clarithromycin in gastric biopsy specimens from dyspeptic patients of a city in the interior of São Paulo, Brazil.BMC Gastroenterol. 2013 Dec 4;13:164. doi: 10.1186/1471-230X-13-164. BMC Gastroenterol. 2013. PMID: 24305035 Free PMC article.
-
Detection of clarithromycin-resistant Helicobacter pylori by stool PCR in children: a comprehensive review of literature.Helicobacter. 2013 Apr;18(2):89-101. doi: 10.1111/hel.12016. Epub 2012 Oct 12. Helicobacter. 2013. PMID: 23067446 Review.
-
Accuracy of Fecal Polymerase Chain Reaction Testing in Clarithromycin-Resistant Helicobacter Pylori: A Systematic Review and Meta-Analysis.Clin Transl Gastroenterol. 2025 Feb 1;16(2):e00792. doi: 10.14309/ctg.0000000000000792. Clin Transl Gastroenterol. 2025. PMID: 39620581 Free PMC article.
Cited by
-
Helicobacter pylori Secondary Antibiotic Resistance after One or More Eradication Failure: A Genotypic Stool Analysis Study.Antibiotics (Basel). 2024 Apr 7;13(4):336. doi: 10.3390/antibiotics13040336. Antibiotics (Basel). 2024. PMID: 38667013 Free PMC article.
-
Helicobacter pylori detection and antimicrobial susceptibility testing.Clin Microbiol Rev. 2007 Apr;20(2):280-322. doi: 10.1128/CMR.00033-06. Clin Microbiol Rev. 2007. PMID: 17428887 Free PMC article. Review.
-
Mechanisms of Helicobacter pylori antibiotic resistance and molecular testing.Front Mol Biosci. 2014 Oct 24;1:19. doi: 10.3389/fmolb.2014.00019. eCollection 2014. Front Mol Biosci. 2014. PMID: 25988160 Free PMC article. Review.
-
Clinical evaluation of a novel H. pylori fecal molecular diagnosis kit (multiplex RT-PCR method) for detecting clarithromycin and fluoroquinolones resistance using stool samples.Front Cell Infect Microbiol. 2025 Jun 23;15:1592612. doi: 10.3389/fcimb.2025.1592612. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40625832 Free PMC article.
-
PNA-FISH as a new diagnostic method for the determination of clarithromycin resistance of Helicobacter pylori.BMC Microbiol. 2011 May 14;11:101. doi: 10.1186/1471-2180-11-101. BMC Microbiol. 2011. PMID: 21569555 Free PMC article.
References
-
- Alarcon, T., A. E. Vega, D. Domingo, M. J. Martinez, and M. Lopez-Brea. 2003. Clarithromycin resistance among Helicobacter pylori strains isolated from children: prevalence and study of mechanism of resistance by PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 41:486-499. - PMC - PubMed
-
- Cadranel, S., L. Corvaglia, P. Bontems, C. Depet, Y. Glupczynski, A. van Riet, and E. Kappens. 1998. Detection of Helicobacter pylori infection in children with a standardized and simplified 13C-urea breath test. J. Pediatr. Gastroenterol. Nutr. 27:275-280. - PubMed
-
- Cavallini A, M. Notarnicola, P. Berloco, A. Lippolis, and A. De Leo. 2000. Use of macroporous polypropylene filter to allow identification of bacteria by PCR in human fecal samples. J. Microbiol. Methods 39:265-270. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

