Prolonged photoresponses and defective adaptation in rods of Gbeta5-/- mice
- PMID: 12904457
- PMCID: PMC6740649
- DOI: 10.1523/JNEUROSCI.23-18-06965.2003
Prolonged photoresponses and defective adaptation in rods of Gbeta5-/- mice
Abstract
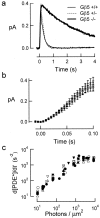
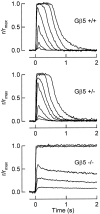
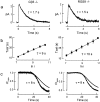
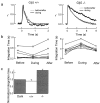
Timely deactivation of G-protein signaling is essential for the proper function of many cells, particularly neurons. Termination of the light response of retinal rods requires GTP hydrolysis by the G-protein transducin, which is catalyzed by a protein complex that includes regulator of G-protein signaling RGS9-1 and the G-protein beta subunit Gbeta5-L. Disruption of the Gbeta5 gene in mice (Gbeta5-/-) abolishes the expression of Gbeta5-L in the retina and also greatly reduces the expression level of RGS9-1. We examined transduction in dark- and light-adapted rods from wild-type and Gbeta5-/- mice. Responses of Gbeta5-/- rods were indistinguishable in all respects from those of RGS9-/- rods. Loss of Gbeta5-L (and RGS9-1) had no effect on the activation of the G-protein cascade, but profoundly slowed its deactivation and interfered with the speeding of incremental dim flashes during light adaptation. Both RGS9-/- and Gbeta5-/- responses were consistent with another factor weakly regulating GTP hydrolysis by transducin in a manner proportional to the inward current. Our results indicate that a complex containing RGS9-1-Gbeta5-L is essential for normal G-protein deactivation and rod function. In addition, our light adaptation studies support the notion than an additional weak GTPase-accelerating factor in rods is regulated by intracellular calcium and/or cGMP.
Figures
Similar articles
-
Slowed recovery of rod photoresponse in mice lacking the GTPase accelerating protein RGS9-1.Nature. 2000 Feb 3;403(6769):557-60. doi: 10.1038/35000601. Nature. 2000. PMID: 10676965
-
Absence of the RGS9.Gbeta5 GTPase-activating complex in photoreceptors of the R9AP knockout mouse.J Biol Chem. 2004 Jan 16;279(3):1581-4. doi: 10.1074/jbc.C300456200. Epub 2003 Nov 18. J Biol Chem. 2004. PMID: 14625292
-
Membrane attachment is key to protecting transducin GTPase-activating complex from intracellular proteolysis in photoreceptors.J Neurosci. 2011 Oct 12;31(41):14660-8. doi: 10.1523/JNEUROSCI.3516-11.2011. J Neurosci. 2011. PMID: 21994382 Free PMC article.
-
The role of Gβ5 in vision.Prog Mol Biol Transl Sci. 2009;86:229-48. doi: 10.1016/S1877-1173(09)86008-0. Epub 2009 Oct 7. Prog Mol Biol Transl Sci. 2009. PMID: 20374718 Review.
-
Phototransduction in mouse rods and cones.Pflugers Arch. 2007 Aug;454(5):805-19. doi: 10.1007/s00424-006-0194-y. Epub 2007 Jan 17. Pflugers Arch. 2007. PMID: 17226052 Free PMC article. Review.
Cited by
-
Transducin translocation in rods is triggered by saturation of the GTPase-activating complex.J Neurosci. 2007 Jan 31;27(5):1151-60. doi: 10.1523/JNEUROSCI.5010-06.2007. J Neurosci. 2007. PMID: 17267570 Free PMC article.
-
Pepperberg plot: Modeling flash response saturation in retinal rods of mouse.Front Mol Neurosci. 2023 Jan 13;15:1054449. doi: 10.3389/fnmol.2022.1054449. eCollection 2022. Front Mol Neurosci. 2023. PMID: 36710929 Free PMC article.
-
Visualizing the chaperone-mediated folding trajectory of the G protein β5 β-propeller.Mol Cell. 2023 Nov 2;83(21):3852-3868.e6. doi: 10.1016/j.molcel.2023.09.032. Epub 2023 Oct 17. Mol Cell. 2023. PMID: 37852256 Free PMC article.
-
Ablation of the GNB3 gene in mice does not affect body weight, metabolism or blood pressure, but causes bradycardia.Cell Signal. 2014 Nov;26(11):2514-20. doi: 10.1016/j.cellsig.2014.07.030. Epub 2014 Aug 2. Cell Signal. 2014. PMID: 25093805 Free PMC article.
-
Novel form of adaptation in mouse retinal rods speeds recovery of phototransduction.J Gen Physiol. 2003 Dec;122(6):703-12. doi: 10.1085/jgp.200308938. Epub 2003 Nov 10. J Gen Physiol. 2003. PMID: 14610022 Free PMC article.
References
-
- Chen C-K, Burns ME, He W, Wensel TG, Baylor DA, Simon MI ( 2000) Slowed recovery of rod photoresponse in mice lacking the GTPase accelerating protein RGS9-1. Nature 403: 557-560. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
