The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense
- PMID: 12904578
- PMCID: PMC187787
- DOI: 10.1073/pnas.1733690100
The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense
Abstract
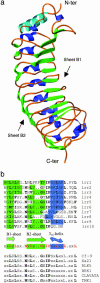
Polygalacturonase-inhibiting proteins (PGIPs) are plant cell wall proteins that protect plants from fungal invasion. They interact with endopolygalacturonases secreted by phytopathogenic fungi, inhibit their enzymatic activity, and favor the accumulation of oligogalacturonides, which activate plant defense responses. PGIPs are members of the leucine-rich repeat (LRR) protein family that in plants play crucial roles in development, defense against pathogens, and recognition of beneficial microbes. Here we report the crystal structure at 1.7-A resolution of a PGIP from Phaseolus vulgaris. The structure is characterized by the presence of two beta-sheets instead of the single one originally predicted by modeling studies. The structure also reveals a negatively charged surface on the LRR concave face, likely involved in binding polygalacturonases. The structural information on PGIP provides a basis for designing more efficient inhibitors for plant protection.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

