Regulation of TCF ETS-domain transcription factors by helix-loop-helix motifs
- PMID: 12907712
- PMCID: PMC169972
- DOI: 10.1093/nar/gkg689
Regulation of TCF ETS-domain transcription factors by helix-loop-helix motifs
Abstract
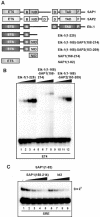
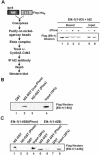
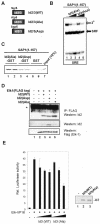
DNA binding by the ternary complex factor (TCF) subfamily of ETS-domain transcription factors is tightly regulated by intramolecular and intermolecular interactions. The helix-loop-helix (HLH)-containing Id proteins are trans-acting negative regulators of DNA binding by the TCFs. In the TCF, SAP-2/Net/ERP, intramolecular inhibition of DNA binding is promoted by the cis-acting NID region that also contains an HLH-like motif. The NID also acts as a transcriptional repression domain. Here, we have studied the role of HLH motifs in regulating DNA binding and transcription by the TCF protein SAP-1 and how Cdk-mediated phosphorylation affects the inhibitory activity of the Id proteins towards the TCFs. We demonstrate that the NID region of SAP-1 is an autoinhibitory motif that acts to inhibit DNA binding and also functions as a transcription repression domain. This region can be functionally replaced by fusion of Id proteins to SAP-1, whereby the Id moiety then acts to repress DNA binding in cis. Phosphorylation of the Ids by cyclin-Cdk complexes results in reduction in protein-protein interactions between the Ids and TCFs and relief of their DNA-binding inhibitory activity. In revealing distinct mechanisms through which HLH motifs modulate the activity of TCFs, our results therefore provide further insight into the role of HLH motifs in regulating TCF function and how the inhibitory properties of the trans-acting Id HLH proteins are themselves regulated by phosphorylation.
Figures




References
-
- Treisman R. (1994) Ternary complex factors: growth regulated transcriptional activators. Curr. Opin. Genet. Dev., 4, 96–101. - PubMed
-
- Sharrocks A.D. (2002) Complexities in ETS-domain transcription factor function and regulation; lessons from the TCF subfamily. Biochem. Soc. Trans., 30, 1–9. - PubMed
-
- Gille H., Sharrocks,A.D. and Shaw,P.E. (1992) Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature, 358, 414–417. - PubMed
-
- Sharrocks A.D. (1995) ERK2/p42 MAP kinase stimulates both autonomous and SRF-dependent DNA binding by Elk-1. FEBS Lett., 368, 77–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

