Alterations in the intracellular level of a protein subunit of human RNase P affect processing of tRNA precursors
- PMID: 12907726
- PMCID: PMC169977
- DOI: 10.1093/nar/gkg691
Alterations in the intracellular level of a protein subunit of human RNase P affect processing of tRNA precursors
Abstract
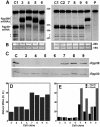
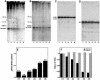
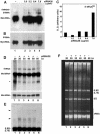
The human ribonucleoprotein ribonuclease P (RNase P), processing tRNA, has at least 10 distinct protein subunits. Many of these subunits, including the autoimmune antigen Rpp38, are shared by RNase MRP, a ribonucleoprotein enzyme required for processing of rRNA. We here show that constitutive expression of exogenous, tagged Rpp38 protein in HeLa cells affects processing of tRNA precursors. Alterations in the site-specific cleavage and in the steady-state level of 3' sequences of the internal transcribed spacer 1 of rRNA are also observed. These processing defects are accompanied by selective shut-off of expression of Rpp38 and by low expression of the tagged protein. RNase P purified from these cells exhibits impaired activity in vitro. Moreover, inhibition of Rpp38 by the use of small interfering RNA causes accumulation of the initiator methionine tRNA precursor. Expression of other protein components, but not of the H1 RNA subunit, is coordinately inhibited. Our results reveal that normal expression of Rpp38 is required for the biosynthesis of intact RNase P and for the normal processing of stable RNA in human cells.
Figures





Similar articles
-
Functional characterization of the conserved amino acids in Pop1p, the largest common protein subunit of yeast RNases P and MRP.RNA. 2006 Jun;12(6):1023-37. doi: 10.1261/rna.23206. Epub 2006 Apr 17. RNA. 2006. PMID: 16618965 Free PMC article.
-
Parallels in rRNA processing: conserved features in the processing of the internal transcribed spacer 1 in the pre-rRNA from Schizosaccharomyces pombe.Biochemistry. 2005 Dec 27;44(51):16977-87. doi: 10.1021/bi051465a. Biochemistry. 2005. PMID: 16363811
-
Localization in the nucleolus and coiled bodies of protein subunits of the ribonucleoprotein ribonuclease P.J Cell Biol. 1999 Aug 9;146(3):559-72. doi: 10.1083/jcb.146.3.559. J Cell Biol. 1999. PMID: 10444065 Free PMC article.
-
Coordination of transcription and processing of tRNA.FEBS J. 2022 Jul;289(13):3630-3641. doi: 10.1111/febs.15904. Epub 2021 May 11. FEBS J. 2022. PMID: 33929081 Review.
-
RNase MRP and rRNA processing.Mol Biol Rep. 1995-1996;22(2-3):69-73. doi: 10.1007/BF00988708. Mol Biol Rep. 1995. PMID: 8901490 Review.
Cited by
-
Heterodimerization regulates RNase MRP/RNase P association, localization, and expression of Rpp20 and Rpp25.RNA. 2007 Jan;13(1):65-75. doi: 10.1261/rna.237807. Epub 2006 Nov 21. RNA. 2007. PMID: 17119099 Free PMC article.
-
Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects.FEBS Lett. 2010 Jan 21;584(2):287-96. doi: 10.1016/j.febslet.2009.11.048. FEBS Lett. 2010. PMID: 19931535 Free PMC article. Review.
-
Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing.J Cell Biol. 2013 Mar 4;200(5):577-88. doi: 10.1083/jcb.201207131. Epub 2013 Feb 25. J Cell Biol. 2013. PMID: 23439679 Free PMC article.
-
MPP6 is an exosome-associated RNA-binding protein involved in 5.8S rRNA maturation.Nucleic Acids Res. 2005 Dec 7;33(21):6795-804. doi: 10.1093/nar/gki982. Print 2005. Nucleic Acids Res. 2005. PMID: 16396833 Free PMC article.
-
A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription.Genes Dev. 2006 Jun 15;20(12):1621-35. doi: 10.1101/gad.386706. Genes Dev. 2006. PMID: 16778078 Free PMC article.

