ADAR2 A-->I editing: site selectivity and editing efficiency are separate events
- PMID: 12907730
- PMCID: PMC169957
- DOI: 10.1093/nar/gkg681
ADAR2 A-->I editing: site selectivity and editing efficiency are separate events
Abstract
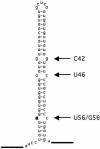
ADAR enzymes, adenosine deaminases that act on RNA, form a family of RNA editing enzymes that convert adenosine to inosine within RNA that is completely or largely double-stranded. Site-selective A-->I editing has been detected at specific sites within a few structured pre-mRNAs of metazoans. We have analyzed the editing selectivity of ADAR enzymes and have chosen to study the naturally edited R/G site in the pre-mRNA of the glutamate receptor subunit B (GluR-B). A comparison of editing by ADAR1 and ADAR2 revealed differences in the specificity of editing. Our results show that ADAR2 selectively edits the R/G site, while ADAR1 edits more promiscuously at several other adenosines in the double-stranded stem. To further understand the mechanism of selective ADAR2 editing we have investigated the importance of internal loops in the RNA substrate. We have found that the immediate structure surrounding the editing site is important. A purine opposite to the editing site has a negative effect on both selectivity and efficiency of editing. More distant internal loops in the substrate were found to have minor effects on site selectivity, while efficiency of editing was found to be influenced. Finally, changes in the RNA structure that affected editing did not alter the binding abilities of ADAR2. Overall these findings suggest that binding and catalysis are independent events.
Figures




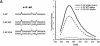
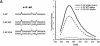
References
-
- Gerber A.P. and Keller,W. (2001) RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem. Sci., 26, 376–384. - PubMed
-
- Burns C.M., Chu,H., Rueter,S.M., Hutchinson,L.K., Canton,H., Sanders-Bush,E. and Emeson,R.B. (1997) Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature, 387, 303–308. - PubMed
-
- Higuchi M., Single,F.N., Kohler,M., Sommer,B., Sprengel,R. and Seeburg,P.H. (1993) RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell, 75, 1361–1370. - PubMed
-
- Lomeli H., Mosbacher,J., Melcher,T., Hoger,T., Geiger,J.R., Kuner,T., Monyer,H., Higuchi,M., Bach,A. and Seeburg,P.H. (1994) Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science, 266, 1709–1713. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

