Characterisation of the substrate specificity of homogeneous vaccinia virus uracil-DNA glycosylase
- PMID: 12907738
- PMCID: PMC169932
- DOI: 10.1093/nar/gkg672
Characterisation of the substrate specificity of homogeneous vaccinia virus uracil-DNA glycosylase
Abstract
The decision to stop smallpox vaccination and the loss of specific immunity in a large proportion of the population could jeopardise world health due to the possibility of a natural or provoked re-emergence of smallpox. Therefore, it is mandatory to improve the current capability to prevent or treat such infections. The DNA repair protein uracil-DNA glycosylase (UNG) is one of the viral enzymes important for poxvirus pathogenesis. Consequently, the inhibition of UNG could be a rational strategy for the treatment of infections with poxviruses. In order to develop inhibitor assays for UNG, as a first step, we have characterised the recombinant vaccinia virus UNG (vUNG) and compared it with the human nuclear form (hUNG2) and catalytic fragment (hUNG) UNG. In contrast to hUNG2, vUNG is strongly inhibited in the presence of 7.5 mM MgCl(2). We have shown that highly purified vUNG is not inhibited by a specific uracil-DNA glycosylase inhibitor. Interestingly, both viral and human enzymes preferentially excise uracil when it is opposite to cytosine. The present study provides the basis for the design of specific inhibitors for vUNG.
Figures
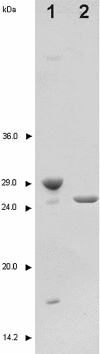
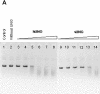


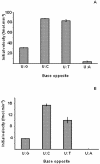

References
-
- Goebel S.J., Johnson,G.P., Perkus,M.E., Davis,S.W., Winslow,J.P. and Paoletti,E. (1990) The complete DNA sequence of vaccinia virus. Virology, 179, 247–266, 517–263. - PubMed
-
- Moss B. (2001) Poxviridae: the virus and their replication. In Knipe,D.M. and Howley,P.M. (eds), Fields Virology. Raven Press, New York, Vol. 2, pp. 2849–2884.
-
- Traktman P. (1990) Poxviruses: an emerging portrait of biological strategy. Cell, 62, 621–626. - PubMed
-
- Moss B., Ahn,B.Y., Amegadzie,B., Gershon,P.D. and Keck,J.G. (1991) Cytoplasmic transcription system encoded by vaccinia virus. J. Biol. Chem., 266, 1355–1358. - PubMed
-
- Traktman P. (1996) Poxvirus DNA replication. In DePamphilis,M.L. (ed.), DNA Replication in Eucaryotic Cells. Cold Spring Harbor Laboratory Press, New York, pp. 775–793.

