Combination of DNA-directed immobilization and immuno-PCR: very sensitive antigen detection by means of self-assembled DNA-protein conjugates
- PMID: 12907742
- PMCID: PMC169982
- DOI: 10.1093/nar/gng090
Combination of DNA-directed immobilization and immuno-PCR: very sensitive antigen detection by means of self-assembled DNA-protein conjugates
Abstract
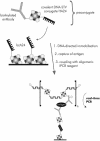
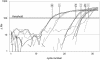
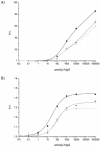
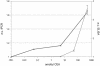
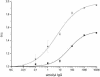
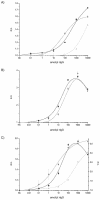
An assay for very sensitive antigen detection is described which takes advantage of the self- assembly capabilities of semi-synthetic conjugates of DNA and proteins. The general scheme of this assay is similar to a two-sided (sandwich) enzyme-linked immunoassay (ELISA); however, covalent single-stranded DNA-streptavidin (STV) conjugates, capable of hybridizing to complementary surface-bound DNA oligomers, are utilized for the effective immobilization of either capture antibodies or antigens, rather than the chemi- or physisorption usually applied in ELISA. Immuno-PCR (IPCR) is employed as a method for signal generation, utilizing oligomeric reagents obtained by self-assembly of STV, biotinylated DNA and antibodies. In three different model systems, detecting human IgG, rabbit IgG or carcinoembryonic antigen, this combination allowed one to increase the sensitivity of the analogous ELISA approximately 1000-fold. For example, <0.1 amol/ micro l (15 pg/ml) of rabbit IgG was detectable. The immunoassay can be carried out in a single step by tagging the analyte with both reagents for capture and read-out simultaneously, thereby significantly reducing handling time and costs of analysis. Moreover, as the spatial selectivity of target immobilization is determined by the specificity of DNA base pairing, the assay is particularly suited for miniaturized microfluidics and lab-on-a-chip devices.
Figures
References
-
- Khandurina J. and Guttman,A. (2002) Bioanalysis in microfluidic devices. J. Chromatogr. A, 943, 159–183. - PubMed
-
- Burke D.T., Burns,M.A. and Mastrangelo,C. (1997) Microfabrication technologies for integrated nucleic acid analysis. Genome Res., 7, 189–197. - PubMed
-
- Quake S.R. and Scherer,A. (2000) From micro- to nanofabrication with soft materials. Science, 290, 1536–1540. - PubMed
-
- Thorsen T., Maerkl,S.J. and Quake,S.R. (2002) Microfluidic large-scale integration. Science, 298, 580–584. - PubMed
-
- Pirrung M.C. (2002) How to make a DNA chip. Angew. Chem. Int. Ed., 41, 1276–1289. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

