The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling
- PMID: 12908874
- PMCID: PMC333403
- DOI: 10.1186/1475-4924-2-20
The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling
Abstract
Background: Forkhead transcription factors belonging to the FOXO subfamily are negatively regulated by protein kinase B (PKB) in response to signaling by insulin and insulin-like growth factor in Caenorhabditis elegans and mammals. In Drosophila, the insulin-signaling pathway regulates the size of cells, organs, and the entire body in response to nutrient availability, by controlling both cell size and cell number. In this study, we present a genetic characterization of dFOXO, the only Drosophila FOXO ortholog.
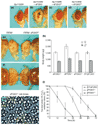
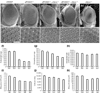
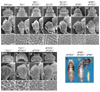
Results: Ectopic expression of dFOXO and human FOXO3a induced organ-size reduction and cell death in a manner dependent on phosphoinositide (PI) 3-kinase and nutrient levels. Surprisingly, flies homozygous for dFOXO null alleles are viable and of normal size. They are, however, more sensitive to oxidative stress. Furthermore, dFOXO function is required for growth inhibition associated with reduced insulin signaling. Loss of dFOXO suppresses the reduction in cell number but not the cell-size reduction elicited by mutations in the insulin-signaling pathway. By microarray analysis and subsequent genetic validation, we have identified d4E-BP, which encodes a translation inhibitor, as a relevant dFOXO target gene.
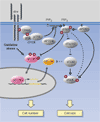
Conclusion: Our results show that dFOXO is a crucial mediator of insulin signaling in Drosophila, mediating the reduction in cell number in insulin-signaling mutants. We propose that in response to cellular stresses, such as nutrient deprivation or increased levels of reactive oxygen species, dFOXO is activated and inhibits growth through the action of target genes such as d4E-BP.
Figures



References
-
- Takahashi Y, Kadowaki H, Momomura K, Fukushima Y, Orban T, Okai T, Taketani Y, Akanuma Y, Yazaki Y, Kadowaki T. A homozygous kinase-defective mutation in the insulin receptor gene in a patient with leprechaunism. Diabetologia. 1997;40:412–420. - PubMed
-
- Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. - PubMed
-
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. - PubMed
-
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. - PubMed
-
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–221. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

