Activation of Notch signaling pathway precedes heart regeneration in zebrafish
- PMID: 12909711
- PMCID: PMC304103
- DOI: 10.1073/pnas.1834204100
Activation of Notch signaling pathway precedes heart regeneration in zebrafish
Abstract
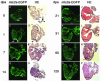
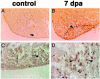
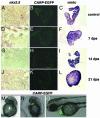
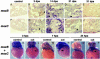
Several vertebrates display the ability to regenerate parts of their body after amputation. During this process, differentiated cells reenter the cell cycle and proliferate to generate a mass of undifferentiated cells. Repatterning mechanisms act on these cells to eventually shape a regenerated tissue or organ that replaces the amputated one. Experiments with regenerating limbs/fins in newts and zebrafish have shown that members of the Msx family of homeodomain-containing transcription factors play key roles during blastema formation and patterning. Here we show that adult zebrafish have a remarkable capacity to regenerate the heart in a process that involves up-regulation of msxB and msxC genes. We present evidence indicating that heart regeneration involves the execution of a specific genetic program, rather than redeployment of a cardiac development program. Preceding Msx activation, there is a marked increase in the expression of notch1b and deltaC, which we show are also up-regulated during fin regeneration. These data suggest a role for the Notch pathway in the activation of the regenerative response. Taken together, our results underscore the use of zebrafish as a model for investigating the process of regeneration in particular and the biology of stem cells in general. Advances in these fields will undoubtedly aid in the implementation of strategies for regenerative medicine.
Figures


References
-
- Tsonis, P. A. (2000) Dev. Biol. 221, 273-284. - PubMed
-
- Brockes, J. P. & Kumar, A. (2002) Nat. Rev. Mol. Cell Biol. 3, 566-574. - PubMed
-
- Brockes, J. P. (1997) Science 276, 81-87. - PubMed
-
- Bryant, S. V., Endo, T. & Gardiner, D. M. (2002) Int. J. Dev. Biol. 46, 887-896. - PubMed
-
- Gardiner, D. M. & Bryant, S. V. (1996) Int. J. Dev. Biol. 40, 797-805. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

