Atypical expansion in mice of the sensory neuron-specific Mrg G protein-coupled receptor family
- PMID: 12909716
- PMCID: PMC187757
- DOI: 10.1073/pnas.1732949100
Atypical expansion in mice of the sensory neuron-specific Mrg G protein-coupled receptor family
Abstract
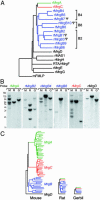
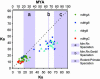
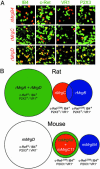
The Mas-related genes (Mrgs) comprise a family of >50 G protein-coupled receptors (GPCRs), many of which are expressed in specific subsets of nociceptive sensory neurons in mice. In contrast, humans contain a related but nonorthologous family of genes, called MrgXs or sensory neuron-specific receptors, of which many fewer appear to be expressed in sensory neurons. To determine whether the diversity of murine Mrgs is generic to rodents or is an atypical feature of mice, we characterized MrgA, MrgB, MrgC, and MrgD subfamilies in rat and gerbil. Surprisingly, although mice have approximately 22 MrgA and approximately 14 MrgC genes, rats and gerbils have just a single MrgA and MrgC gene. This murine-specific expansion likely reflects recent retrotransposon-mediated unequal crossover events. The expression of Mrgs in rat sensory ganglia suggests that the extensive cellular diversity in mice can be simplified to a core subset of approximately four different genes (MrgA, MrgB, MrgC, and MrgD), defining a similar number of neuronal subpopulations. Our results suggest more generally that mouse-human genomic comparisons may sometimes reveal differences atypical of rodents.
Figures


References
-
- Young, J. M. & Trask, B. J. (2002) Hum. Mol. Genet. 11, 1153–1160. - PubMed
-
- Lindemann, B. (2001) Nature 413, 219–225. - PubMed
-
- Nathans, J. (1999) Neuron 24, 299–312. - PubMed
-
- Wang, Y., Macke, J. P., Merbs, S. L., Zack, D. J., Klaunberg, B., Bennett, J., Gearhart, J. & Nathans, J. (1992) Neuron 9, 429–440. - PubMed
-
- Vassalli, A., Rothman, A., Feinstein, P., Zapotocky, M. & Mombaerts, P. (2002) Neuron 35, 681–696. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

