Sequence elements in both the intergenic space and the 3' untranslated region of the Crithidia fasciculata KAP3 gene are required for cell cycle regulation of KAP3 mRNA
- PMID: 12912886
- PMCID: PMC178339
- DOI: 10.1128/EC.2.4.671-677.2003
Sequence elements in both the intergenic space and the 3' untranslated region of the Crithidia fasciculata KAP3 gene are required for cell cycle regulation of KAP3 mRNA
Abstract
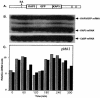
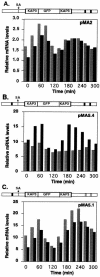
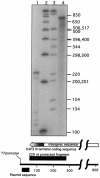
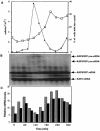
mRNA levels of several Crithidia fasciculata genes involved in DNA metabolism have previously been found to cycle as cells progress through the cell cycle. Octamer consensus sequences in the 5' untranslated regions (5' UTRs) of these transcripts were shown to be required for cycling of these mRNAs. The KAP3 gene encodes a kinetoplast histone H1-like DNA binding protein, and its mRNA levels cycle in parallel with those of the kinetoplast DNA topoisomerase (TOP2), dihydrofolate reductase-thymidylate synthase (DHFR-TS), and the large subunit of the nuclear single-stranded DNA binding protein (RPA1). KAP3 mRNA contains two octamer consensus sequences in its 3' UTR but none in its 5' UTR. Mutation of these octamer sequences was not sufficient to prevent cycling of a sequence-tagged KAP3 mRNA expressed from a plasmid. Mutation of an octamer sequence contained on the precursor transcript but not on the mRNA, in addition to mutation of the two octamer sequences in the 3' UTR, was necessary to abolish cycling of the mRNA. The requirement for a sequence not present on the mature mRNA indicates that regulation of the mRNA levels by the octamer sequences occurs at or prior to splicing of the transcript. Incompletely processed RNAs containing octamer sequences were also found to accumulate during the cell cycle when the mRNA levels were lowest. These RNA species hybridize to both the KAP3 coding sequence and that of the downstream drug resistance gene, indicating a lack of processing within the intergenic region separating these genes. We propose a cell cycle-dependent interference in transcript processing mediated by octamer consensus sequences as a mechanism contributing to the cycling of such transcripts.
Figures


References
-
- Black, D. L. 1991. Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev. 5:389-402. - PubMed
-
- Di Noia, J. M., I. D'Orso, D. O. Sanchez, and A. C. Frasch. 2000. AU-rich elements in the 3′-untranslated region of a new mucin-type gene family of Trypanosoma cruzi confers mRNA instability and modulates translation efficiency. J. Biol. Chem. 275:10218-10227. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

