Inositol and phosphate regulate GIT1 transcription and glycerophosphoinositol incorporation in Saccharomyces cerevisiae
- PMID: 12912892
- PMCID: PMC178388
- DOI: 10.1128/EC.2.4.729-736.2003
Inositol and phosphate regulate GIT1 transcription and glycerophosphoinositol incorporation in Saccharomyces cerevisiae
Erratum in
- Eukaryot Cell. 2003 Dec;2(6):1386
Abstract
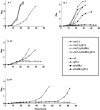
Glycerophosphoinositol is produced through deacylation of the essential phospholipid phosphatidylinositol. In Saccharomyces cerevisiae, the glycerophosphoinositol produced is excreted from the cell but is recycled for phosphatidylinositol synthesis when inositol is limiting. To be recycled, glycerophosphoinositol enters the cell through the permease encoded by GIT1. The transport of exogenous glycerophosphoinositol through Git1p is sufficiently robust to support the growth of an inositol auxotroph (ino1Delta). We now report that S. cerevisiae also uses exogenous phosphatidylinositol as an inositol source. Evidence suggests that phosphatidylinositol is deacylated to glycerophosphoinositol extracellularly before being transported across the plasma membrane by Git1p. A genetic screen identified Pho86p, which is required for targeting of the major phosphate transporter (Pho84p) to the plasma membrane, as affecting the utilization of phosphatidylinositol and glycerophosphoinositol. Deletion of PHO86 in an ino1Delta strain resulted in faster growth when either phosphatidylinositol or glycerophosphoinositol was supplied as the sole inositol source. The incorporation of radiolabeled glycerophosphoinositol into an ino1Delta pho86Delta mutant was higher than that into wild-type, ino1Delta, and pho86Delta strains. All strains accumulated the most GIT1 transcript when incubated in media limited for inositol and phosphate in combination. However, the ino1Delta pho86Delta mutant accumulated approximately threefold more GIT1 transcript than did the other strains when incubated in inositol-free media containing either high or low concentrations of P(i). Deletion of PHO4 abolished GIT1 transcription in a wild-type strain. These results indicate that the transport of glycerophosphoinositol by Git1p is regulated by factors affecting both inositol and phosphate availabilities and suggest a regulatory connection between phosphate metabolism and phospholipid metabolism.
Figures


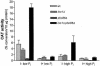
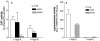
Similar articles
-
Glycerophosphoinositol, a novel phosphate source whose transport is regulated by multiple factors in Saccharomyces cerevisiae.J Biol Chem. 2004 Jul 23;279(30):31937-42. doi: 10.1074/jbc.M403648200. Epub 2004 May 15. J Biol Chem. 2004. PMID: 15145930
-
GIT1, a gene encoding a novel transporter for glycerophosphoinositol in Saccharomyces cerevisiae.Genetics. 1998 Aug;149(4):1707-15. doi: 10.1093/genetics/149.4.1707. Genetics. 1998. PMID: 9691030 Free PMC article.
-
Posttranscriptional regulation of Git1p, the glycerophosphoinositol/glycerophosphocholine transporter of Saccharomyces cerevisiae.Curr Genet. 2006 Dec;50(6):367-75. doi: 10.1007/s00294-006-0096-8. Epub 2006 Aug 22. Curr Genet. 2006. PMID: 16924500
-
Transport and metabolism of glycerophosphodiesters produced through phospholipid deacylation.Biochim Biophys Acta. 2007 Mar;1771(3):337-42. doi: 10.1016/j.bbalip.2006.04.013. Epub 2006 May 6. Biochim Biophys Acta. 2007. PMID: 16781190 Review.
-
Messengers for morphogenesis: inositol polyphosphate signaling and yeast pseudohyphal growth.Curr Genet. 2019 Feb;65(1):119-125. doi: 10.1007/s00294-018-0874-0. Epub 2018 Aug 12. Curr Genet. 2019. PMID: 30101372 Review.
Cited by
-
Identification of genes affecting vacuole membrane fragmentation in Saccharomyces cerevisiae.PLoS One. 2013;8(2):e54160. doi: 10.1371/journal.pone.0054160. Epub 2013 Feb 1. PLoS One. 2013. PMID: 23383298 Free PMC article.
-
The glycerophosphocholine acyltransferase Gpc1 is part of a phosphatidylcholine (PC)-remodeling pathway that alters PC species in yeast.J Biol Chem. 2019 Jan 25;294(4):1189-1201. doi: 10.1074/jbc.RA118.005232. Epub 2018 Dec 4. J Biol Chem. 2019. PMID: 30514764 Free PMC article.
-
Phospholipid turnover and acyl chain remodeling in the yeast ER.Biochim Biophys Acta Mol Cell Biol Lipids. 2020 Jan;1865(1):158462. doi: 10.1016/j.bbalip.2019.05.006. Epub 2019 May 27. Biochim Biophys Acta Mol Cell Biol Lipids. 2020. PMID: 31146038 Free PMC article. Review.
-
Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast.J Biol Chem. 2005 Mar 11;280(10):9106-18. doi: 10.1074/jbc.M411770200. Epub 2004 Dec 20. J Biol Chem. 2005. PMID: 15611057 Free PMC article.
-
Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae.Curr Genet. 2010 Feb;56(1):1-32. doi: 10.1007/s00294-009-0287-1. Curr Genet. 2010. PMID: 20054690 Review.
References
-
- Angus, W. W., and R. L. Lester. 1972. Turnover of inositol and phosphorus containing lipids in Saccharomyces cerevisiae; extracellular accumulation of glycerophosphorylinositol derived from phosphatidylinositol. Arch. Biochem. Biophys. 151:483-495. - PubMed
-
- Bachhawat, N., Q. Ouyang, and S. A. Henry. 1995. Functional characterization of an inositol-sensitive upstream activation sequence in yeast: a cis-regulatory element responsible for inositol-choline mediated regulation of phospholipid biosynthesis. J. Biol. Chem. 270:25087-25095. - PubMed
-
- Bun-ya, M., K. Shikata, S. Nakade, C. Yompakdee, S. Harashima, and Y. Oshima. 1996. Two new genes, PHO86 and PHO87, involved in inorganic phosphate uptake in Saccharomyces cerevisiae. Curr. Genet. 29:344-351. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

