Gain-of-function mutations indicate that Escherichia coli Kch forms a functional K+ conduit in vivo
- PMID: 12912904
- PMCID: PMC175798
- DOI: 10.1093/emboj/cdg409
Gain-of-function mutations indicate that Escherichia coli Kch forms a functional K+ conduit in vivo
Abstract
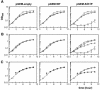
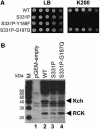
Although Kch of Escherichia coli is thought to be a K(+) channel by sequence homology, there is little evidence that it actually conducts K(+) ions in vitro or in vivo. We isolated gain-of-function (GOF) Kch mutations that render bacteria specifically sensitive to K(+) ions. Millimolar added K(+), but not Na(+) or sorbitol, blocks the initiation or continuation of mutant growth in liquid media. The mutations are mapped at the RCK (or KTN) domain, which is considered to be the cytoplasmic sensor controlling the gate. Additional mutations directed to the K(+)-filter sequence rescue the GOF mutant. The apparent K(+)-specific conduction through the 'loose-cannon' mutant channel suggests that the wild-type Kch channel also conducts, albeit in a regulated manner. Changing the internal ATG does not erase the GOF toxicity, but removes kch's short second product, suggesting that it is not required for channel function in vivo. The mutant phenotypes are better explained by a perturbation of membrane potential instead of internal K(+) concentration. Possible implications on the normal function of Kch are discussed.
Figures


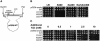

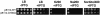

Similar articles
-
The projection structure of Kch, a putative potassium channel in Escherichia coli, by electron crystallography.Biochim Biophys Acta. 2014 Jan;1838(1 Pt B):237-43. doi: 10.1016/j.bbamem.2013.09.006. Epub 2013 Sep 19. Biochim Biophys Acta. 2014. PMID: 24055821
-
Functional properties of Kch, a prokaryotic homologue of eukaryotic potassium channels.Biochem Biophys Res Commun. 2002 Sep 13;297(1):10-6. doi: 10.1016/s0006-291x(02)02095-8. Biochem Biophys Res Commun. 2002. PMID: 12220501
-
An Escherichia coli homologue of eukaryotic potassium channel proteins.Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3510-4. doi: 10.1073/pnas.91.9.3510. Proc Natl Acad Sci U S A. 1994. PMID: 8170937 Free PMC article.
-
Assembly of Kch, a putative potassium channel from Escherichia coli.J Struct Biol. 2009 Nov;168(2):288-93. doi: 10.1016/j.jsb.2009.07.018. Epub 2009 Jul 23. J Struct Biol. 2009. PMID: 19631752
-
Sodium permeability and sensitivity induced by mutations in the selectivity filter of the KcsA channel towards Kir channels.Biochimie. 2010 Mar;92(3):232-44. doi: 10.1016/j.biochi.2009.11.007. Epub 2009 Dec 3. Biochimie. 2010. PMID: 19962419
Cited by
-
A novel manganese efflux system, YebN, is required for virulence by Xanthomonas oryzae pv. oryzae.PLoS One. 2011;6(7):e21983. doi: 10.1371/journal.pone.0021983. Epub 2011 Jul 14. PLoS One. 2011. PMID: 21789199 Free PMC article.
-
Unappreciated Roles for K+ Channels in Bacterial Physiology.Trends Microbiol. 2021 Oct;29(10):942-950. doi: 10.1016/j.tim.2020.11.005. Epub 2020 Dec 5. Trends Microbiol. 2021. PMID: 33288383 Free PMC article. Review.
-
Electrical Impedance Spectroscopy with Bacterial Biofilms: Neuronal-like Behavior.Nano Lett. 2024 Feb 21;24(7):2234-2241. doi: 10.1021/acs.nanolett.3c04446. Epub 2024 Feb 6. Nano Lett. 2024. PMID: 38320294 Free PMC article.
-
Functional characterization and determination of the physiological role of a calcium-dependent potassium channel from cyanobacteria.Plant Physiol. 2013 Jun;162(2):953-64. doi: 10.1104/pp.113.215129. Epub 2013 May 2. Plant Physiol. 2013. PMID: 23640756 Free PMC article.
-
The desensitization gating of the MthK K+ channel is governed by its cytoplasmic amino terminus.PLoS Biol. 2008 Oct 28;6(10):e223. doi: 10.1371/journal.pbio.0060223. PLoS Biol. 2008. PMID: 18959476 Free PMC article.
References
-
- Doyle D.A., Cabral,J.M., Pfuetzner,R.A., Kuo,A.L., Gulbis,J.M., Cohen,S.L., Chait,B.T. and MacKinnon,R. (1998) The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science, 280, 69–77. - PubMed
-
- Hille B. (2001) Ion Channels of Excitable Membranes. Sinauer Associates, Sunderland, MA.
-
- Hirsch R.E., Lewis,B.D., Spalding,E.P. and Sussman,M.R. (1998) A role for the AKT1 potassium channel in plant nutrition. Science, 280, 918–921. - PubMed
-
- Jiang Y.X., Pico,A., Cadene,M., Chait,B.T. and MacKinnon,R. (2001) Structure of the RCK domain from the E.coli K+ channel and demonstration of its presence in the human BK channel. Neuron, 29, 593–601. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

