Interdimer processing mechanism of procaspase-8 activation
- PMID: 12912912
- PMCID: PMC175803
- DOI: 10.1093/emboj/cdg414
Interdimer processing mechanism of procaspase-8 activation
Abstract
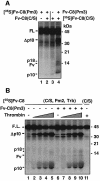
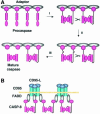
The execution of apoptosis depends on the hierarchical activation of caspases. The initiator procaspases become autoproteolytically activated through a less understood process that is triggered by oligomerization. Procaspase-8, an initiator caspase recruited to death receptors, is activated through two cleavage events that proceed in a defined order to generate the large and small subunits of the mature protease. Here we show that dimerization of procaspase-8 produces enzymatically competent precursors through the stable homophilic interaction of the procaspase-8 protease domain. These dimers are also more susceptible to processing than individual procaspase-8 molecules, which leads to their cross-cleavage. The order of the two interdimer cleavage events is maintained by a sequential accessibility mechanism: the separation of the large and small subunits renders the region between the large subunit and prodomain susceptible to further cleavage. In addition, the activation process involves an alteration in the enzymatic properties of caspase-8; while procaspase-8 molecules specifically process one another, mature caspases only cleave effector caspases. These results reveal the key steps leading to the activation of procaspase-8 by oligomerization.
Figures





References
-
- Ashkenazi A. and Dixit,V.M. (1998) Death receptors: signaling and modulation. Science, 281, 1305–1308. - PubMed
-
- Blanchard H., Donepudi,M., Tschopp,M., Kodandapani,L., Wu,J.C. and Grutter,M.G. (2000) Caspase-8 specificity probed at subsite S(4): crystal structure of the caspase-8-Z-DEVD-CHO complex. J. Mol. Biol., 302, 9–16. - PubMed
-
- Boatright K.M. et al. (2003) A unified model for apical caspase activation. Mol. Cell, 11, 529–541. - PubMed
-
- Chang D.W. and Yang,X. (2003) Activation of procaspases by FK506 binding protein-mediated oligomerization. Sci. STKE, 2003, PL1. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

