A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs
- PMID: 12912925
- PMCID: PMC175784
- DOI: 10.1093/emboj/cdg394
A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs
Abstract
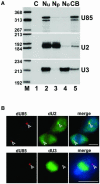
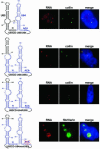
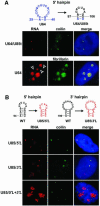
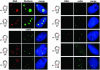
Post-transcriptional synthesis of 2'-O-methylated nucleotides and pseudouridines in Sm spliceosomal small nuclear RNAs takes place in the nucleoplasmic Cajal bodies and it is directed by guide RNAs (scaRNAs) that are structurally and functionally indistinguishable from small nucleolar RNAs (snoRNAs) directing rRNA modification in the nucleolus. The scaRNAs are synthesized in the nucleoplasm and specifically targeted to Cajal bodies. Here, mutational analysis of the human U85 box C/D-H/ACA scaRNA, followed by in situ localization, demonstrates that box H/ACA scaRNAs share a common Cajal body-specific localization signal, the CAB box. Two copies of the evolutionarily conserved CAB consensus (UGAG) are located in the terminal loops of the 5' and 3' hairpins of the box H/ACA domains of mammalian, Drosophila and plant scaRNAs. Upon alteration of the CAB boxes, mutant scaRNAs accumulate in the nucleolus. In turn, authentic snoRNAs can be targeted into Cajal bodies by addition of exogenous CAB box motifs. Our results indicate that scaRNAs represent an ancient group of small nuclear RNAs which are localized to Cajal bodies by an evolutionarily conserved mechanism.
Figures


References
-
- Bachellerie J.P., Cavaille,J. and Qu,L.H. (2000) Nucleotide modification of eukaryotic rRNAs: the world of small nucleolar RNAs revisited. In Garrett,R.A., Douthwaite,S.R., Matheson,A.T., Moure,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 191–203.
-
- Bachellerie J.P., Cavaille,J. and Huttenhofer,A. (2002) The expanding snoRNA world. Biochimie, 84, 775–790. - PubMed
-
- Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell, 86, 823–834. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

