Protein grafting of an HIV-1-inhibiting epitope
- PMID: 12913122
- PMCID: PMC187838
- DOI: 10.1073/pnas.1733910100
Protein grafting of an HIV-1-inhibiting epitope
Abstract
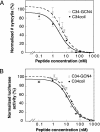
Protein grafting, the transfer of a binding epitope of one ligand onto the surface of another protein, is a potentially powerful technique for presenting peptides in preformed and active three-dimensional conformations. Its utility, however, has been limited by low biological activity of the designed ligands and low tolerance of the protein scaffolds to surface substitutions. Here, we graft the complete binding epitope (19 nonconsecutive amino acids with a solvent-accessible surface area of >2,000 A2) of an HIV-1 C-peptide, which is derived from the C-terminal region of HIV-1 gp41 and potently inhibits HIV-1 entry into cells, onto the surface of a GCN4 leucine zipper. The designed peptide, named C34coil, displays a potent antiviral activity approaching that of the native ligand. Moreover, whereas the linear C-peptide is unstructured and sensitive to degradation by proteases, C34coil is well structured, conformationally stable, and exhibits increased resistance to proteolytic degradation compared with the linear peptide. In addition to being a structured antiviral inhibitor, C34coil may also serve as the basis for the development of an alternative class of immunogens. This study demonstrates that "one-shot" protein grafting, without subsequent rounds of optimization, can be used to create ligands with structural conformations and improved biomedical properties.
Figures



References
-
- D'Souza, M. P., Cairns, J. S. & Plaeger, S. F. (2000) J. Am. Med. Assoc. 284, 215–222. - PubMed
-
- Eckert, D. M. & Kim, P. S. (2001) Annu. Rev. Biochem. 70, 777–810. - PubMed
-
- Moore, J. P. & Stevenson, M. (2000) Nat. Rev. Mol. Cell Biol. 1, 40–49. - PubMed
-
- Nabel, G. J. (2001) Nature 410, 1002–1007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

