Overexpression of the disease resistance gene Pto in tomato induces gene expression changes similar to immune responses in human and fruitfly
- PMID: 12913147
- PMCID: PMC181276
- DOI: 10.1104/pp.103.022731
Overexpression of the disease resistance gene Pto in tomato induces gene expression changes similar to immune responses in human and fruitfly
Abstract
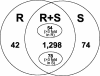
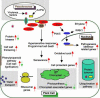
The Pto gene encodes a serine/threonine protein kinase that confers resistance in tomato (Lycopersicon esculentum) to Pseudomonas syringae pv tomato strains that express the type III effector protein AvrPto. Constitutive overexpression of Pto in tomato, in the absence of AvrPto, activates defense responses and confers resistance to several diverse bacterial and fungal plant pathogens. We have used a series of gene discovery and expression profiling methods to examine the effect of Pto overexpression in tomato leaves. Analysis of the tomato expressed sequence tag database and suppression subtractive hybridization identified 600 genes that were potentially differentially expressed in Pto-overexpressing tomato plants compared with a sibling line lacking Pto. By using cDNA microarrays, we verified changes in expression of many of these genes at various time points after inoculation with P. syringae pv tomato (avrPto) of the resistant Pto-overexpressing line and the susceptible sibling line. The combination of these three approaches led to the identification of 223 POR (Pto overexpression responsive) genes. Strikingly, 40% of the genes induced in the Pto-overexpressing plants previously have been shown to be differentially expressed during the human (Homo sapiens) and/or fruitfly (Drosophila melanogaster) immune responses.
Figures


References
-
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JDG, Parker JE (2002) Regulatory role of SGT1 in Early R gene-mediated plant defenses. Science 295: 2077–2080 - PubMed
-
- Axelrod AE (1981) Role of the B vitamins in the immune response. Adv Exp Med Biol 135: 93–106 - PubMed
-
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295: 2073–2076 - PubMed
-
- Beck MA (2001) Antioxidants and viral infections: host immune response and viral pathogenicity. J Am Coll Nutr 20: 384S–388S - PubMed
-
- Ben-Neriah Y (2002) Regulatory functions of ubiquitination in the immune system. Nat Immunol 3: 20–26 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

