Comparative genomics of Archaea: how much have we learned in six years, and what's next?
- PMID: 12914651
- PMCID: PMC193635
- DOI: 10.1186/gb-2003-4-8-115
Comparative genomics of Archaea: how much have we learned in six years, and what's next?
Abstract
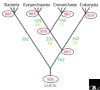
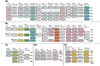
Archaea comprise one of the three distinct domains of life (with bacteria and eukaryotes). With 16 complete archaeal genomes sequenced to date, comparative genomics has revealed a conserved core of 313 genes that are represented in all sequenced archaeal genomes, plus a variable 'shell' that is prone to lineage-specific gene loss and horizontal gene exchange. The majority of archaeal genes have not been experimentally characterized, but novel functional pathways have been predicted.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

