Refinement of a chimpanzee pericentric inversion breakpoint to a segmental duplication cluster
- PMID: 12914658
- PMCID: PMC193642
- DOI: 10.1186/gb-2003-4-8-r50
Refinement of a chimpanzee pericentric inversion breakpoint to a segmental duplication cluster
Abstract
Background: Pericentric inversions are the most common euchromatic chromosomal differences among humans and the great apes. The human and chimpanzee karyotype differs by nine such events, in addition to several constitutive heterochromatic increases and one chromosomal fusion event. Reproductive isolation and subsequent speciation are thought to be the potential result of pericentric inversions, as reproductive boundaries form as a result of hybrid sterility.
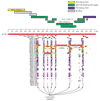
Results: Here we employed a comparative fluorescence in situ hybridization approach, using probes selected from a combination of physical mapping, genomic sequence, and segmental duplication analyses to narrow the breakpoint interval of a pericentric inversion in chimpanzee involving the orthologous human 15q11-q13 region. We have refined the inversion breakpoint of this chimpanzee-specific rearrangement to a 600 kilobase (kb) interval of the human genome consisting of entirely duplicated material. Detailed analysis of the underlying sequence indicated that this region comprises multiple segmental duplications, including a previously characterized duplication of the alpha7 neuronal nicotinic acetylcholine receptor subunit gene (CHRNA7) in 15q13.3 and several Golgin-linked-to-PML, or LCR15, duplications.
Conclusions: We conclude that, on the basis of experimental data excluding the CHRNA7 duplicon as the site of inversion, and sequence analysis of regional duplications, the most likely rearrangement site is within a GLP/LCR15 duplicon. This study further exemplifies the genomic plasticity due to the presence of segmental duplications and highlights their importance for a complete understanding of genome evolution.
Figures


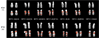

References
-
- Yunis JJ, Sawyer JR, Dunham K. The striking resemblance of high-resolution G-banded chromosomes of man and chimpanzee. Science. 1980;208:1145–1148. - PubMed
-
- Yunis JJ, Prakash O. The origin of man: a chromosomal pictorial legacy. Science. 1982;215:1525–1530. - PubMed
-
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. - PubMed
-
- Dehal P, Predki P, Olsen AS, Kobayashi A, Folta P, Lucas S, Land M, Terry A, Ecale Zhou CL, Rash S, et al. Human chromosome 19 and related regions in mouse: conservative and lineage-specific evolution. Science. 2001;293:104–111. - PubMed
-
- Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

